2788
Repeatability of ultra-high-resolution Multi-Parametric Mapping across five 7T sites1GIGA-Cyclotron Research Centre - In Vivo Imaging, University of Liège, Liège, Belgium, 2MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom, 3Wellcome Centre for Human Neuroimaging, University College London, London, United Kingdom, 4Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 5Wolfson Brain Imaging Centre, University of Cambridge, Cambridge, United Kingdom, 6Spinal cord injury Center, University of Zurich, Zurich, Switzerland, 7Swiss Center for Musculoskeletal Imaging, Zurich, Switzerland, 8University of Zurich, Zurich, Switzerland, 9Department of Neurology, University Hospital of Liège, Liège, Belgium, 10Felix Bloch Institute for Solid State Physics, Leipzig University, Leipzig, Germany, 11GIGA - In Silico Medicine, University of Liège, Liège, Belgium
Synopsis
Repeatability is key to the utility of quantitative MRI, which promises standardised measures with biological relevance. Here we tested a candidate ultra-high resolution (0.6mm isotropic) multi-parameter mapping protocol at five 7T sites. Repeatability was good, with B0 and B1+ field inhomogeneities being the limiting factor. MTsat had the lowest repeatability and PD the highest. R1 and R2* were intermediate. Repeatability was also lowest in temporal and occipital cortices. These observations were consistent across sites.
INTRODUCTION
Quantitative MRI provides access to markers of biologically-relevant tissue features and offers a standardised approach to imaging that can improve comparability across sites and time points 1. This study sought to assess the repeatability of an ultra-high resolution (0.6mm isotropic) quantitative Multi-Parameter Mapping (MPM) protocol across five different 7T MR imaging sites.METHODS
A healthy volunteer (M, 34Y) underwent two MPM sessions with half an hour break in between (“scan” and “rescan”) on a Siemens MAGNETOM Terra, running VE12U-SP01 software, equipped with a single transmit, 32 channel receive head coil (Nova Medical). Imaging was repeated with the same volunteer, within 4 weeks, at five sites: University College London, UK (UCL), University of Cambridge, UK (CAM), Swiss Center for Musculoskeletal Imaging, Balgrist Campus, Switzerland (BAL), Max Planck Institute for Human Cognitive and Brain Sciences, Germany (CBS) and the University of Liege, Belgium (ULG)).Automatic calibration, with three iterations, was used to set the transmitter reference voltage. This was followed by whole-brain B0 shimming (WIP1441), the outcome of which was used for all subsequent scans. The MPM protocol consisted of three 3D multi-echo gradient-echo scans with T1 (flip angle, FA = 200), PD (FA = 50) or MT weighting (4ms long Gaussian pulse, 2kHz off-resonance with a FA of 1400)2,3. Additional parameters: TR 19.50ms, 6 equidistant echoes with TE ranging from 2.3 to 14.2ms (reduced to 4 for MT-weighting), bandwidth of 469Hz/pixel, RF spoiling increment of 1440 with 4pi dephasing per TR4, non-selective rectangular excitation pulse with a constant duration of 240μs across flip angles, GRAPPA factor 2 with 48 integrated reference lines in each phase-encoded direction, and a total acquisition time of 9 minutes 10 seconds per volume. The transmit field, B1+, was mapped using a series of spin-echo and stimulated echo images (SESTE)5,6 or via the Bloch-Siegert shift (BSS)7.
MPMs (MTsat, PD, R1, R2*) were calculated using the hMRI toolbox3. This was repeated for each B1 mapping method (only SESTE available from CAM). To assess repeatability, the intra-site coefficient of variation (CoV) was calculated as the difference relative to the mean on a voxel-wise basis expressed in percentage. Regions-of-interest (ROIs) for quantitative analysis were defined by merging the cortical and subcortical Harvard-Oxford atlases into 65 (anatomically defined) regions8. These were transformed to native space and restricted to voxels with a GM probability >80% via masking. The median CoV was extracted from each ROI.
RESULTS
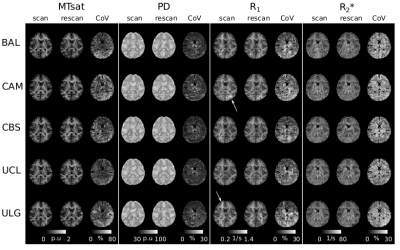
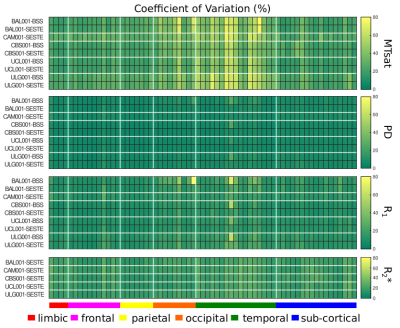
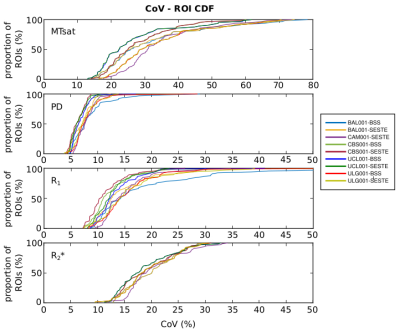
Figure-1 shows a single plane of each map from the scan and rescan sessions together with its CoV for each site. The quality of the parameter maps is generally good, however, distributed artefacts are visible in the R1 maps (e.g. Fig.1 arrows and discernible inter-hemispheric asymmetries).Figure 2 shows the regional variation of the CoV. The highest CoV, indicating lower repeatability, values were observed for the MTsat map and within the temporal lobe. These findings were consistent across sites. Figure 3 shows the cumulative density of the median CoV values across ROIs and sites. 90% of ROIs had CoV values (median(± IQR) across sites) below 51.33(± 9.44)%, 9.78(±1.76)%, 21.73 (±6.06)% and 26.61(±0.31)% for MTsat, PD, R1 and R2*respectively.
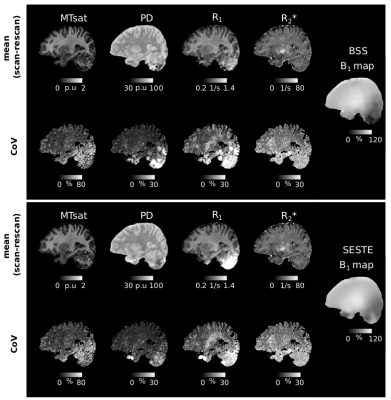
B1+ mapping was particularly challenging in the cerebellum due to low B1+ efficiency and high off-resonance, more so for the SESTE method. Figure 4 shows the mean MPMs (across scan and rescan) for one site (BAL) for both BSS and SESTE, together with the corresponding plane from the B1+ maps. The cerebellar region was very noisy and gave inconsistent or infeasible values, most visibly in the R1 map. Hence, the cerebellum was excluded from the CoV analysis. Low B1+ efficiency also leads to higher CoVs in some parts of the temporal and occipital lobes.
DISCUSSION
The repeatability of the MPM measures was generally good and consistent across sites and the results were in line with the CoV values reported for high-resolution (0.8mm) MPMs at 3T9 but higher for MTsat, which had the lowest repeatability. PD was most reproducible while R1 and R2* were intermediate. Spatially, repeatability was lowest in temporal and occipital cortices.Distributed artefacts were observed in the R1 maps. These may be caused by intra-scan motion, though no obvious motion artefacts were visible in the FLASH volumes. Inter-scan motion can also lead to substantial variation in transmit and receive fields at 7T and may have contributed to spatially varying error in R1. Off-resonance conditions are challenging for the FLASH volumes and particularly the B1+ mapping techniques used. SESTE uses an EPI readout with low resolution making it vulnerable to distortions and intra-voxel dephasing. BSS uses a matched FLASH readout but is phase-based and contaminated by higher-order B0 inhomogeneity effects. These effects reduced the repeatability (e.g. temporal lobe, Figure 2) and led to substantial artefacts in the cerebellum (Figure 4).
CONCLUSION
This study assessed the repeatability of an ultra-high-resolution (0.6mm) MPM protocol by scanning the same individual twice at 5 different 7T sites. The repeatability of the maps was good and consistent at all sites. Further investigations are required to more fully evaluate the inter-scanner and inter-individual variability. This study can serve as a baseline for the assessment of other protocols.Acknowledgements
The authors would like to acknowledge Siemens Healthineers for the support of our project through the provision of the WIP sequence used for B0 mapping and B0 shimming (WIP 1441). CTR is funded by a Sir Henry Dale Fellowship from the Wellcome Trust and the Royal Society (#098436/Z/12/B). We acknowledge support from the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. MFC The Wellcome Centre for Human Neuroimaging is supported by core funding from the Wellcome [203147/Z/16/Z]. SS, EB, GV, CP, CB is supported by ULiège-Valeo Innovation Chair, Siemens Healthineers, FNRS-Belgium, European Regional Development Fund (Radiomed project). NW was supported by the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n° 616905, the European Union's Horizon 2020 research and innovation programme under the grant agreement No 681094, the BMBF (01EW1711A & B) in the framework of ERA-NET NEURON. BD is funded by the Gates Cambridge Scholarship. PF is funded by an SNF Eccellenza Professorial Fellowship grant (PCEFP3_181362 / 1) and received a grant from EMDO Foundation (No. 1100). The CRC has an institutional agreement with Siemens Healthineers and benefits from logistic support. SS is funded by Siemens healthineers. SS and GV received funds from Valeo. AA is an employee of Siemens Healthcare. The Max Planck Institute for Human Cognitive and Brain Sciences has an institutional research agreement with Siemens Healthcare. NW holds a patent on acquisition of MRI data during spoiler gradients (US 10,401,453 B2). NW was a speaker at an event organized by Siemens Healthcare and was reimbursed for the travel expenses.References
1. Weiskopf N, Edwards LJ, Helms G, Mohammadi S, Kirilina E. Quantitative magnetic resonance imaging of brain anatomy and in vivo histology. Nature Reviews Physics. 2021;3(8):570-588.
2. Weiskopf N, Suckling J, Williams G, et al. Quantitative multi-parameter mapping of R1, PD(*), MT, and R2(*) at 3T: a multi-center validation. Front Neurosci. 2013;7:95.
3. Tabelow K, Balteau E, Ashburner J, et al. hMRI - A toolbox for quantitative MRI in neuroscience and clinical research. Neuroimage. 2019;194:191-210.
4. Corbin N, Callaghan MF. Imperfect spoiling in variable flip angle T 1 mapping at 7T: Quantifying and minimizing impact. Magnetic Resonance in Medicine. 2021;86(2):693-708.
5. Lutti A, Hutton C, Finsterbusch J, Helms G, Weiskopf N. Optimization and validation of methods for mapping of the radiofrequency transmit field at 3T. Magn Reson Med. 2010;64(1):229-238.
6. Lutti A, Stadler J, Josephs O, et al. Robust and fast whole brain mapping of the RF transmit field B1 at 7T. PLoS One. 2012;7(3):e32379.
7. Corbin N, Acosta-Cabronero J, Malik SJ, Callaghan MF. Robust 3D Bloch-Siegert based B 1 + mapping using multi-echo general linear modeling. Magn Reson Med. 2019;82(6):2003-2015.
8. Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968-980.
9. Callaghan MF, Josephs O, Herbst M, Zaitsev M, Todd N, Weiskopf N. An evaluation of prospective motion correction (PMC) for high resolution quantitative MRI. Front Neurosci. 2015;9:97.
Figures