2784
PySynthMRI: An open-source Python tool for Synthetic MRI
Luca Peretti1,2,3, Matteo Cencini3, Paolo Cecchi2,4, Graziella Donatelli2,4, Mauro Costagli3,5, and Michela Tosetti3
1University of Pisa, Pisa, Italy, 2Imago7 Research Foundation, Pisa, Italy, 3IRCCS Stella Maris, Pisa, Italy, 4Azienda Ospedaliero-Universitaria Pisana, Pisa, Italy, 5University of Genoa, Genoa, Italy
1University of Pisa, Pisa, Italy, 2Imago7 Research Foundation, Pisa, Italy, 3IRCCS Stella Maris, Pisa, Italy, 4Azienda Ospedaliero-Universitaria Pisana, Pisa, Italy, 5University of Genoa, Genoa, Italy
Synopsis
Synthetic MR imaging allows reconstruction of different image contrasts from a single acquisition, reducing scan times. Here we introduce PySynthMRI, an open-source tool which provides a user-friendly and fluid interface to synthesize and modify contrast-weighted images. The tool allows the user to add and adjust virtual scanner parameters showing, in real time, how the resulting synthesized images vary accordingly, which makes PySynthMRI a valuable tool for both research and teaching. The generated images can be exported in the preferred format.
Introduction
MRI is a noninvasive modality which allows one to acquire images at high spatial resolution and it produces excellent soft tissue contrasts. A complete MRI exam typically consists of several sequences, designed to obtain images with different contrast This leads to long acquisition time, which can be challenging for children and uncollaborative patients. In this scenario, a method to synthesize MR images with adjustable parameters is desirable.Current approaches synthesize image contrasts based on measurements of tissue parameters from a single acquisition1, reducing the scanning time. This technique, known as Synthetic MRI, allows to obtain images by post processing acquired data and radiologists can retrospectively manipulate the image to change the contrasts. MAGnetic resonance image Compilation (MAGiC)2 and SyMRI3 are examples of products that implement synthetic MRI. Those tools, capable of generating and visualizing synthetic images, are vendor-specific and we are not aware of open-source tools to achieve the same results using in-house developed multiparametric imaging techniques.
We introduce PySynthMRI, an open-source tool that offers a friendly interface (Figure 1) that uses quantitative maps of T1, T2 and proton density (PD) as input and allows the user to vary scanner settings such as echo time (TE), repetition time (TR), inversion time (TI), that mimic those used in conventional scans. Using the signal equations for different types of acquisitions, the tool dynamically synthesizes and shows to the user the contrast images, such as T1 weighted, T2 weighted, FLAIR . The simplicity of use makes PySynthMRI a valuable tool for both research and teaching.
Design and features
The PySynthMRI tool is written in Python 3.6, an open programming language available in all computing platforms. The tool makes use of the open-source numpy, open-cv and pyqtgraph packages, chosen with the aim of maximizing the efficiency in manipulating image matrices and therefore to make the interaction with the graphical interface fluid for the user. The code is organized in a Model-View-Controller (MVC) pattern and its modularity simplifies the tests and makes it possible to add new features without impacting the codebase.The interface of PySynthMRI allows the user to load the needed quantitative maps, including but not limited to T1, T2 and proton density quantitative maps. It provides a simple way to modify sequence parameters in order to choose values that generate the synthetic image, which is immediately shown to the user using the preferred interpolation for best rendering.
New custom sequence parameters, including their weights in the signal equation, can be added using configuration files or dynamically in order to obtain unconventional contrast weighted images. PySynthMRI generates the images S as a function of chosen scanner parameters and provided quantitative maps, according to the model represented by
$$abs(\prod_{p\ \in\ params} o_p+s_p*e^{\frac{-p}{Q_p}})$$
where each scanner parameter p is associated to a different quantitative map Q, an offset o and a scale factor s.
Since scanner parameters are independent, any contrast image can be obtained. To facilitate the user, the tool contains a set of default parameters in its configuration file used to synthesize conventional contrast images. T1-weighted MP2RAGE (Magnetization-Prepared 2 RApid Gradient Echoes) and GRE (Gradient Echo) are obtained respectively using TI and TR.
$$MP2RAGE =abs(1 - 2 * exp(-TI /T1)))$$
$$GRE =abs(1 - 2 * exp(-TR /T1))$$
The T2-weighted FSE (Fast Spin Echo) image is synthesized using the echo time, scaled on proton density as follows
$$FSE =abs(PD *exp(-TE/T2))$$
Furthermore, FLAIR (Fluid Attenuated Inversion Recovery) is obtained according to the following equation
$$FLAIR =abs(PD *exp(-TE/T2)*exp(-T1SAT/T1)*(1 - 2 * exp(-TI /T1)))$$
Once generated, the synthetic images can be exported in the preferred format, obtaining a new file with updated metadata.
The tool is in continuous extension. The source code as well as documentation is released on GitHub under free MIT software license and can be accessed under: https://github.com/FiRMLAB-Pisa/pySynthMRI.git
Discussion and Conclusion
The development and dissemination of quantitative MRI techniques such as Magnetic Resonance Fingerprinting makes the PySynthMRI tool beneficial for both MRI research and teaching. In the research field the graphical interface of our tool allows specialists to easily synthesize all needed contrast images at post-processing time, granting the possibility to reach the optimal contrast weighting for that particular subject. In teaching it can be used to show how sequence parameters impact the generation of images with different tissue contrast.The modular architecture allows developers and contributors to easily extend and customize the tool to incorporate additional parameter maps such as T2* or MT or different predefined contrast images. The tool functionality could be further improved including a segmentation software module to immediately show how different generated images impact on the computed results.
Acknowledgements
No acknowledgement found.References
- Gómez PA, Cencini M, Golbabaee M, et al. Rapid three-dimensional multiparametric MRI with quantitative transient-state imaging. Sci Rep. 2020;10(1):1-17. doi:10.1038/s41598-020-70789-2
- Tanenbaum LN, Tsiouris AJ, Johnson AN, et al. Synthetic MRI for clinical neuroimaging: results of the magnetic resonance image compilation (MAGiC) prospective, multicenter, multireader trial. AJNR Am J Neuroradiol. 2017;38:1103–1110. doi: 10.3174/ajnr.A5227
- Blystad I, Warntjes JB, Smedby O, et al. Synthetic MRI of the brain in a clinical setting. Acta Radiol. 2012 Dec 1;53(10):1158-63. doi: 10.1258/ar.2012.120195. PMID: 23024181.
Figures
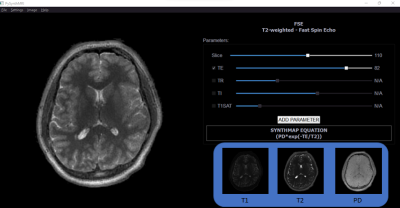
Figure 1. GUI of PySynthMRI. On the left, the synthesized FSE. On the right, the scanner parameters, the resulting image equation and the provided quantitative maps. Navigation bar allows the user to change visual settings such as image orientation and interpolation, switch to different weighted images, add new custom scanner parameters.
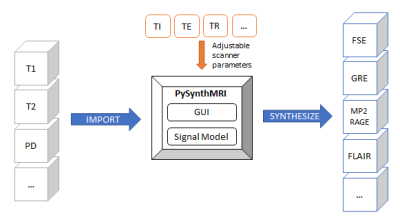
Figure 2. Schematic representation of PySynthMRI basic functionality. The user can import quantitative maps and use the interface to set up scanner parameters. The tool generates synthetic contrast-weighted images at runtime using the signal model.
DOI: https://doi.org/10.58530/2022/2784