2781
A BIDS extension proposal for magnetic resonance spectroscopy data1Department of Radiology, Weill Cornell Medicine, New York, NY, United States, 2Schulich School of Medicine and Dentistry, Western University, London, ON, Canada, 3High-field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 4Department of Psychological and Brain Sciences, Dartmouth College, Hanover, NH, United States, 5Departments of Psychiatry, Neuroscience, and Biomedical Engineering, University of Florida, Gainesville, FL, United States, 6Center for OCD, Anxiety, and Related Disorders, University of Florida, Gainesville, FL, United States, 7Center for Cognitive Aging and Memory, University of Florida, Gainesville, FL, United States, 8McKnight Brain Institute, University of Florida, Gainesville, FL, United States, 9Brain Rehabilitation Research Center, Malcom Randall Veterans Affairs Medical Center, Gainesville, FL, United States, 10Bangor Imaging Unit, School of Psychology, Bangor University, Bangor, United Kingdom, 11Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 12F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 13Centre for Human Brain Health and School of Psychology, University of Birmingham, Birmingham, United Kingdom
Synopsis
Magnetic resonance spectroscopy (MRS) studies can generate extensive and complex datasets. Datasets can be organized idiosyncratically across (and even within) labs. However, sharing data and reproducing results without the added challenge of parsing lab-specific file organization is paramount to open science. Sharing of MRS data that is independent of lab-specific practices would greatly aid transparency and reusability. Here, we present an extension to the Brain Imaging Data Structure (BIDS) specification for MRS data. MRS-BIDS adopts the established tenets of standardization of BIDS and applies them to MRS data while considering the nuances relevant to MRS methodology.
Introduction
Magnetic resonance spectroscopy (MRS) studies can generate extensive and complex datasets, especially when multiple modalities are employed. The organization of such data usually differs between (or even within) labs. This makes the sharing or reusing of data difficult, impeding transparency and reproducibility. To address this, the neuroimaging community created the Brain Imaging Data Structure (BIDS)1 specification (https://bids.neuroimaging.io/). Inspired by the practices of the OpenNeuro repository2, BIDS has served as a standard for the organization of neuroimaging data and metadata, but its principles have also been applied to other biomedical imaging and non-imaging modalities. BIDS currently covers many brain imaging methods, such as MRI techniques (e.g., fMRI, diffusion imaging, and arterial spin labeling3), magnetoencephalography4, electroencephalography5, and positron emission tomography6. Together with ongoing efforts to include other modalities, we propose an extension to BIDS for MRS data, MRS-BID.Summary of BIDS
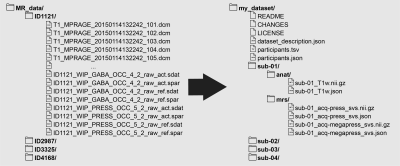
The purpose of BIDS is to reorganize arbitrary data structures into a template that can be more easily used, reused, and parsed (Fig. 1). Since neuroimaging data can be stored in many formats, BIDS facilitates accessibility and interoperability by adopting open-source and standardized file formats, such as JavaScript Object Notation (JSON) and tab-separated values (TSV) files. MR data, in turn, are stored in NIfTI format. At the root of each dataset’s directory tree, there must be files that describe the dataset (README, CHANGES, LICENSE, dataset_description.json) and participants (participants.tsv, participants.json). File name structures follow a chain of key/value pairs. For instance, a data file for a participant from a specific session must begin with the chain sub-<label>_ses-<label>. Many other key/value pairs are possible, and chains are arranged to ensure that every data file and derived output can be identified unambiguously, even if files are misplaced in another directory. A complete description of the current version of the BIDS specification is available online: https://bids-specification.readthedocs.io/.MRS-BIDS specification
The motivation for the MRS-BIDS extension has arisen in part from a push for standardization and sharing of data and software tools in the MRS community in recent years. The formation of the Committee for MRS Code and Data Sharing of the ISMRM’s MR Spectroscopy Study Group is one such example of this. MRS-BIDS adopts the common principles of BIDS while relying on current community consensus recommendations7,8 for MRS-specific data and metadata considerations.Full details on the MRS-BIDS specification can be found online (https://bit.ly/3ATZzwz); a general description of the proposal as it stands is provided below. Furthermore, we wish to emphasize that this extension proposal is, and must be, community-driven. We encourage community members to contribute to the online document and provide their feedback on the MRSHub Forum (https://bit.ly/31qWXu5). This extension is a work-in-progress and will continue to evolve.
Data file formats
MRS data files vary considerably, with each MRI manufacturer having one or more proprietary formats. We believe it is imperative to adopt a single file format standard for MRS-BIDS. The recently created open-source NIfTI-MRS data format9 is an ideal choice. Like the native NIfTI format for MRI data, NIfTI-MRS makes it easy to load raw data and parse the header for relevant metadata.Directory structure
In MRS-BIDS, NIfTI-MRS data must be stored within an mrs/ directory under each participant sub-<label>/ directory. File naming largely follows the BIDS templates used for MRI data. At a minimum, file names must begin and end with sub-<label> and <suffix>.nii.gz, respectively, where the suffix would either be svs or mrsi to indicate the data are single-voxel spectroscopy or MRS imaging acquisitions. To accommodate different acquisition parameters, BIDS already offers solutions. For example, in TE-averaging acquisitions where echo time is varied, the key/value pair echo-<index> can be used to distinguish between files. Data files from inversion recovery experiments to null metabolite signals can use inv-<index>. If there are multiple scans acquired using different pulse sequences, such as PRESS and MEGA-PRESS, the acq-<label> could be used to distinguish the two (or more) data files (see Fig. 1).Sidecar JSON files
Each NIfTI-MRS file must have an associated sidecar JSON file (*_<suffix>.json) storing all relevant metadata that describes the data. This sidecar should contain common metadata fields shared with other MR modalities (e.g., Manufacturer and MagneticFieldStrength). Required, recommended, and optional MRS-specific fields (e.g., ResonantNucleus, SpectrometerFrequency, and VoxelSize) are too numerous to list here and can be found online (https://bit.ly/3ATZzwz).Derivatives
Derivative data (e.g., voxel masks and tissue segmentation maps) would follow the same file naming structure already defined by BIDS for derivative imaging data. As with raw data, sidecar JSON files must also be present.Conclusion
MRS-BIDS offers a standard for organizing MRS data. BIDS is widely adopted by the neuroimaging community, making the addition of an MRS data specification highly appropriate. We hope that MRS software developers will take full advantage of the benefits of MRS-BIDS and adapt their tools to be BIDS-compatible. Given the straightforward guidelines of BIDS, it is envisioned that it will become increasingly easier to automate data analysis pipelines and derive reproducible output across labs using MRS.Acknowledgements
We thank all the individuals who have contributed to this extension proposal. We also wish to thank the BIDS maintainers (https://bit.ly/3C5gf4P) for providing valuable support and direction. MM is supported by NIH grant K99EB028828.References
1. Gorgolewski KJ, Auer T, Calhoun VD, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data. Jun 21 2016;3:160044. doi:10.1038/sdata.2016.44
2. Markiewicz CJ, Gorgolewski KJ, Feingold F, et al. The OpenNeuro resource for sharing of neuroscience data. eLife. 2021;10doi:10.7554/elife.71774
3. Clement P, Castellaro M, Okell TW, et al. ASL-BIDS, the brain imaging data structure extension for arterial spin labeling. PsyArXiv. 2021;doi:10.31234/osf.io/e87y3
4. Niso G, Gorgolewski KJ, Bock E, et al. MEG-BIDS, the brain imaging data structure extended to magnetoencephalography. Sci Data. Jun 19 2018;5:180110. doi:10.1038/sdata.2018.110
5. Pernet CR, Appelhoff S, Gorgolewski KJ, et al. EEG-BIDS, an extension to the brain imaging data structure for electroencephalography. Sci Data. Jun 25 2019;6(1):103. doi:10.1038/s41597-019-0104-8
6. Knudsen GM, Ganz M, Appelhoff S, et al. Guidelines for the content and format of PET brain data in publications and archives: A consensus paper. J Cereb Blood Flow Metab. Aug 2020;40(8):1576-1585. doi:10.1177/0271678X20905433
7. Lin A, Andronesi O, Bogner W, et al. Minimum Reporting Standards for in vivo Magnetic Resonance Spectroscopy (MRSinMRS): Experts' consensus recommendations. NMR Biomed. May 2021;34(5):e4484. doi:10.1002/nbm.4484
8. Kreis R, Boer V, Choi IY, et al. Terminology and concepts for the characterization of in vivo MR spectroscopy methods and MR spectra: Background and experts' consensus recommendations. NMR Biomed. Aug 17 2020:e4347. doi:10.1002/nbm.4347
9. Clark WT, Wilson M. NIfTI-MRS format specification. Zenodo. 2021;doi:10.5281/zenodo.5657097
Figures