2778
Ginkgo: a novel modular and and Open Source MRI pulse sequence development framework dedicated to MRI systems1BAOBAB, NeuroSpin, Paris-Saclay University, CNRS, CEA, Gif-sur-Yvette, France, 2Beauval Nature, ZooParc de Beauval, Saint-Aignan, France, 3UMR 85 Physiologie de la Reproduction et des Comportements, INRAE Centre Val-de-Loire, Nouzilly, France, 4Parietal, NeuroSpin, Paris-Saclay University, Inria, CEA, Gif-sur-Yvette, France
Synopsis
We created a new object-oriented environment for MR pulse-sequence development based on IDEA VE11C and above versions using an Open Science philosophy. This Ginkgo toolkit uses a modular structure to facilitate the design of pulse sequences using the aggregation of basic open-source sequence blocks available from the toolkit. Proofs of concept of the productivity gain reached using Ginkgo are provided through the implementation of a series of sequence models including a diffusion-weighted PGSE 3D EPI sequence.
Introduction
The development of MRI sequences1,2 generally relies on the access to a development kit provided by MRI manufacturers giving access to some proprietary sequence codes from which new sequence concepts are established. Although this approach allows MR physicists to carry out their developments, it remains under the direct control of the provider and often depends on its capacity to open up the codes of which he has the intellectual property. Such limitations have sometimes motivated the scientific community to become autonomous in order to develop ad-hoc solutions, as for example in the case of MRI data reconstruction with the Gadgetron project3. In this work, we propose to adopt a similar approach through the development of an OpenSource MRI sequence development environment, Ginkgo, dedicated to MR systems operating under IDEA (release VE11C and above), and we provide examples of sequences developed using this novel framework.Material and methods
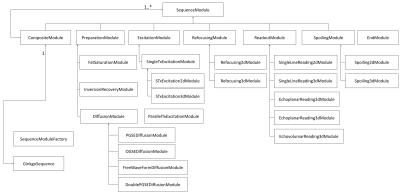
A MRI sequence can be seen as an assembly of various modules (excitation, readout, saturation, inversion, refocusing, diffusion, spoiling, etc.) and described as an ordered sequence of these modules over time, with temporal constraints generally linked to the duration of the modules and their temporal anchoring in the sequence. This concept of module can therefore be associated with a generic SequenceModule class written in C++ language which specifies the hardware limits (maximum gradient magnitude and minimum gradient rise time) that the module cannot exceed and which defines its temporal constraints materialised in the form of a temporal anchor, a total duration and the distribution of this duration before and after the temporal anchor. This class has four methods:- the initialize() method, which allows the operator's prescriptions to be retrieved from the control console,
- the prepare() method, which allows the module to be initialized, to be described and to prepare the various events that make it up (gradient pulses, radio frequency pulses, opening controls of the analogue to digital converters, oscillator control),
- the modifyKernel() method, which allows the amplitude of the gradient pulses that make up the module to be controlled according to the various slice, partition and phase indices in real time, the reading bandwidth to be adjusted if necessary, and the opening of the converters and oscillators to be controlled accordingly,
- the setKernelTiming() method, which allows the module's anchor to be set in the sequence.
Several elementary modules have been developed in the Ginkgo framework by deriving from this parent class to provide the basic elements required to build any MRI sequence as described in the UML diagram on Figure 1. Any module can have knowledge of the other modules to adapt its parameters according to the parameter settings of other modules. A further CompositeModule class allows composition from elementary modules, thus facilitating the construction of MRI sequences from reusable composite modules.
Results
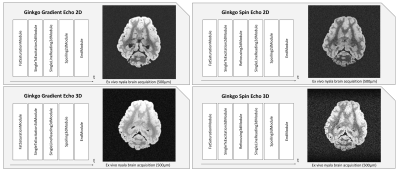
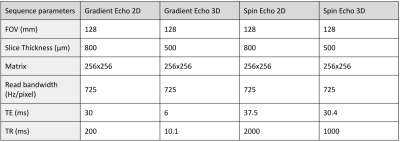
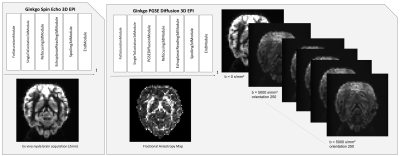
Figure 2 illustrates concrete implementations of basic 2D and 3D gradient echo and spin echo sequences using the different sequence modules available from the Ginkgo toolbox. The sequences were used on a Siemens Prisma 3T MRI system running under VE11C IDEA release to scan an ex vivo nyala brain provided by the ZooParc de Beauval. Sequence parameters were chosen as summarized in Figure 3.Figure 4 shows that the implementation of more complex sequences such as a 3D echoplanar (EPI) spin echo sequence could also be developed efficiently using the Ginkgo toolbox. A diffusion-weighted pulsed-gradient spin echo (PGSE)4,5,6 3D EPI sequence was also developed to assess the modularity of the framework, from which relevant quantitative high angular resolution diffusion imaging (HARDI) maps could be inferred. All EPI sequences were compatible with the use of partial Fourier sampling and parallel acquisition strategies such as GRAPPA to reduce the echo time and scan durations.
Discussion
The Ginkgo toolkit provides a flexible and easy to understand architecture which makes the development of sequences similar to building a set of legos, thanks to the use of a wide range of open-source MRI sequence modules available from its library. We demonstrated its efficacy to create simple sequence schemes as well as more complex ones like 3D diffusion-weighted EPI. It has the potential to allow the design of many innovative sequences, based on the basic and optional modules, as well as on community-proposed modules. Its open-science philosophy frees it from the manufacturer’s limits regarding intellectual property and will allow the research community to contribute to its development. However, it should be noted that it remains dedicated to the Siemens users community having access to MRI systems working under VE11C or above.Conclusion
Following an open science strategy, we have proposed in this work a new object-oriented and modular software environment for the agile development of MRI sequences. The Ginkgo framework , freely available at https://framagit.org/cpoupon/gkg/pulse-sequence, is dedicated to the community of MRI physicists working on Siemens MR systems and aiming at exploiting this novel open source sequence development kit for innovative research or aiming at contributing to its development.Acknowledgements
This work has been partially funded by the Phare PhD Program of the CEA (France).References
1. Bernstein M. A., King K. F., and Xiaohong J. Z.: Handbook of MRI pulse sequences. Elsevier, 2004.
2. Brown R. W. et al. Magnetic resonance imaging: physical principles and sequence design. John Wiley & Sons, 2014.
3. Hansen, M. S. and Sørensen, T. S.: Gadgetron: an open source framework for medical image reconstruction. Magnetic resonance in medicine, 2013; 69(6):1768-1776.
4. Le Bihan D., Breton E., Lallemand D., Grenier P., Cabanis E. and Laval-Jeantet M.: MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology, 1986; 161(2):401-407.
5. Stejskal E. O. and Tanner J. E.: Spin diffusion measurements: spin echoes in the presence of a time‐dependent field gradient. The journal of chemical physics, 1965; 42(1):288-292.
6. Poser B. A., Koopmans P. J., Witzel T., Wald L. L. and Barth M.: Three dimensional echo-planar imaging at 7 Tesla. Neuroimage, 2010; 51(1):261-266.
Figures