2773
A Cloud-based MR Spectroscopy Tool for Basis Set Simulation1Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland School of Medicine, Baltimore, MD, United States, 4Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 5AnatomyWorks, LLC, Ellicott City, MD, United States, 6Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada, 7Sunnybrook Research Institute, Toronto, ON, Canada
Synopsis
MRSCloud is an online-based spectral simulation tool for brain metabolites. Up to 32 metabolites can be simulated to generate a basis set for linear combination modeling. The 1D projection method, coherence pathway filters and pre-calculation of propagators have been implemented for density-matrix simulations to run on a high-performance cloud server. Results indicated simulations were comparable with FID-A and difference between simulations of spatial points (21x21/41x41/101x101) were small. It allows community users to generate vendor, sequence, editing experimental-specific basis sets that are appropriate for their studies.
Introduction
Magnetic resonance spectroscopy (MRS) is a non-invasive method that measures the in vivo concentration of brain metabolites. Accurate quantification requires modeling of acquired spectra using a linear combination of known metabolite basis functions. Basis sets are most commonly generated by numerical simulation because preparation of a full set of metabolite phantoms and experimental acquisition is time-consuming, expensive and extremely demanding technically. Simulations based on the quantum mechanical density-matrix formalism (1) can be carried out in a number of application tools and software packages (2-8).There is a significant learning curve to all these tools, and only a minority of groups applying MRS have the detailed sequence knowledge to implement accurate basis-set simulations locally. Therefore, most analyses rely upon basis sets generated off-site, often with approximate, rather than exact-match parameters. Furthermore, spectral editing sequences have recently been added to product for the major vendors. Users now require basis sets both for conventional and scans edited for J-coupled metabolites, such as GABA, and for more advanced Hadamard-encoded sequences (9,10). This abstract introduces MRSCloud, a cloud-based spectral simulation tool, which allows the user to generate density-matrix-simulated basis sets through a simple web interface. Parameters specified include localization method (PRESS or semi-LASER), vendor (GE, Philips, Siemens), sequence (unedited, MEGA, HERMES, HERCULES), metabolite list, spatial resolution, echo time (TE) and editing pulse frequencies.Methods
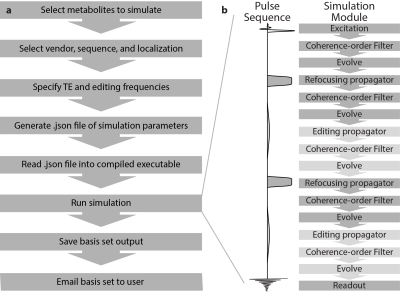
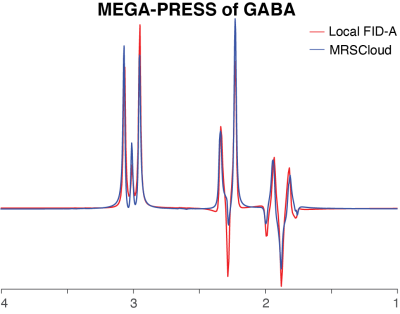
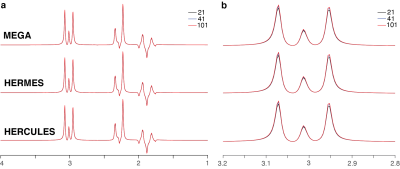
MRSCloud, the GUI for which is shown in Figure 1, is built upon FID-A functionality. Up to 32 metabolites can be simulated, of which 25 use FID-A spin-system definitions (2), augmented by other metabolites including cystathionine, ethanolamine, homocarnosine, lysine, phosphorylethanolamine, threonine and valine (11-13). Other input parameters include localization method (PRESS or semi-LASER), sequence options (Unedited, MEGA, HERMES or HERCULES (9,10,14)), and vendor (GE, Philips and Siemens). TE can be defined for Unedited and MEGA simulations, and is fixed at 80 ms for HERMES and HERCULES. Editing frequencies of ON and OFF scans are user defined for MEGA (default edit-on/-off: 1.9/7.5 ppm) and fixed for HERCULES (1.9/4.18/4.58 ppm) and GABA/GSH-edited HERMES (1.9/4.56 ppm).It has recently been demonstrated in MARSS (5) that density-matrix simulations can be substantially accelerated by using the 1D projection method (15) and applying coherence pathway filters (16,17). MRSCloud includes these functionalities, and makes additional savings by pre-calculating the propagators for all RF pulses. Cloud-based simulations allow exact simulation of proprietary vendor-specific RF pulse shapes, e.g. GTST (18) slice-selective refocusing pulses (bandwidth 1.3 kHz, duration 6.90 ms) for Philips PRESS sequences. GOIA pulses (bandwidth 10 kHz, duration 4.5 ms) are simulated for sLASER localization. Editing pulse durations for MEGA sequences are determined from TE, using a vendor-appropriate shape. A spatial array of simulations (of 101x101 resolution, by default) is carried out across a field of view that extends 50% larger than the voxel size in the two dimensions defined by refocusing pulses.All code was written in MATLAB (R2020b, MathWorks, Natick, USA), and compiled as an executable file to run on the server with MATLAB Runtime. User inputs are stored in a declaration file (.json) that is loaded into the simulation. The pipeline for generating a basis set and the simulation module are shown in Figure 2.For validation, simulations of GABA using MEGA, HERMES and HERCULES at TE 80 ms with PRESS in 21x21, 41x41 and 101x101 spatial points were performed using MRSCloud. For comparison, MEGA-PRESS of GABA in 41x41 spatial points was simulated using the local FID-A.Results
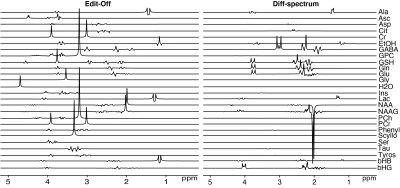
MRSCloud has been successfully established, offering web-based basis set simulation for the community through the https://braingps.anatomyworks.org/mrs-sim portal. Simulation time varies somewhat depending on the usage of the third-party server. MEGA-PRESS simulations of GABA using MRSCloud are comparable to FID-A simulations, as shown in Figure 3, with a run time that is reduced by a factor of 9. MEGA-PRESS, HERMES and HERCULES simulations on GABA at 21x21, 41x41 and 101x101 spatial points are shown in Figure 4. GABA integrals at 3 ppm were slightly larger with a larger number of spatial points. Simulation time of 101x101 spatial points for GABA, a strongly coupled 6-spin system, was less than 1 minute for all three sequences and MEGA-PRESS simulations for a basis set of 25 common metabolites, as shown in Figure 5, was ~4 minutes.Discussion
MRSCloud provides fast and accurate simulation for MRS metabolites and generation of basis sets for linear-combination modeling. This improves access to basis sets across the community, and should result in more modeling being performed with basis sets that exactly represent the timing and RF pulse shapes of specific vendor sequences. Substantial reductions in runtime have been achieved by implementing the 1D projection method, coherence-order filtering, and pre-calculation of propagators, as well as implementation on a high-performance cloud server.Discrepancies in lineshape between MRSCloud and FID-A are driven by the better coherence transfer pathway selection offered by direct filtering of the density matrix, as compared to incomplete phase-cycling of refocusing pulses. This advantage comes in addition to the 4x acceleration of not repeating simulations for the phase cycle. Changes of signal amplitude as the spatial resolution of the simulation changes are largely driven by raster effects (with a larger/smaller fraction of simulated points within the voxel).Acknowledgements
This work was supported by NIH grants R01 EB016089, R01 EB023963, R21 AG060245, R00 AG062230, K99 DA051315, P41 EB015909 and P41 EB031771References
1. Fano U. Description of States in Quantum Mechanics by Density Matrix and Operator Techniques. Reviews of Modern Physics 1957;29(1):74-93.
2. Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J. Advanced processing and simulation of MRS data using the FID appliance (FID-A)-An open source, MATLAB-based toolkit. Magn Reson Med 2017;77(1):23-33.
3. Young K, Matson GB, Govindaraju V, Maudsley AA. Spectral simulations incorporating gradient coherence selection. J Magn Reson 1999;140(1):146-152.
4. Naressi A, Couturier C, Castang I, de Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med 2001;31(4):269-286.
5. Landheer K, Swanberg KM, Juchem C. Magnetic resonance Spectrum simulator (MARSS), a novel software package for fast and computationally efficient basis set simulation. NMR Biomed 2021;34(5):e4129.
6. Bak M, Rasmussen JT, Nielsen NC. SIMPSON: a general simulation program for solid-state NMR spectroscopy. J Magn Reson 2000;147(2):296-330.
7. Veshtort M, Griffin RG. SPINEVOLUTION: a powerful tool for the simulation of solid and liquid state NMR experiments. J Magn Reson 2006;178(2):248-282.
8. Soher B, Semanchuk P, Todd D, Steinberg J, Young K. Vespa: Integrated applications for RF pulse design, spectral simulation and MRS data analysis. 19th Meeting ISMRM. Montreal; 2011.
9. Chan KL, Puts NA, Schar M, Barker PB, Edden RA. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn Reson Med 2016;76(1):11-19.
10. Oeltzschner G, Saleh MG, Rimbault D, et al. Advanced Hadamard-encoded editing of seven low-concentration brain metabolites: Principles of HERCULES. Neuroimage 2019;185:181-190.
11. Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed 2000;13(3):129-153.
12. Branzoli F, Pontoizeau C, Tchara L, et al. Cystathionine as a marker for 1p/19q codeleted gliomas by in vivo magnetic resonance spectroscopy. Neuro Oncol 2019;21(6):765-774.
13. de Graaf RA. In Vivo NMR Spectroscopy – Static Aspects. In Vivo NMR Spectroscopy; 2019. p. 43-128.
14. Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed 1998;11(6):266-272.
15. Zhang Y, An L, Shen J. Fast computation of full density matrix of multispin systems for spatially localized in vivo magnetic resonance spectroscopy. Med Phys 2017;44(8):4169-4178.
16. Mitschang L, Ponstingl H, Grindrod D, Oschkinat H. Geometrical representation of coherence transfer selection by pulsed field gradients in high-resolution nuclear magnetic resonance. The Journal of Chemical Physics 1995;102(8):3089-3098.
17. Bodenhausen G, Kogler H, Ernst RR. Selection of coherence-transfer pathways in NMR pulse experiments. 1984. J Magn Reson 2011;213(2):276-294.
18. Murdoch JB, Lent AH, Kritzer MR. Computer-optimized narrowband pulses for multislice imaging. Journal of Magnetic Resonance (1969) 1987;74(2):226-263.
Figures