2749
Machine Learning based Differentiation of Non-Enhancing Tumor from Vasogenic Edema in patients with Low Grade Gliomas using DCE-MRI1Center for Biomedical Engineering, Indian Institute of Technology, Delhi, India, 2Fortis Memorial Research Institute, Gurugram, India, 3Electrical Engineering, Indian Institute of Technology, Delhi, India, 4Biomedical Engineering, All India Institute of Medical Sciences, Delhi, India
Synopsis
Precise safe surgical resection and precisely directed radiation are important in clinical practice for low-grade gliomas (LGGs) patients in order to minimize the neurological deficit and radiation toxicity, respectively. Clinicians find difficulty in defining the border between the non-enhancing tumor and edema components. In this study, machine learning-based models were developed in order to distinguish non-enhancing tumors from vasogenic edema using quantitative perfusion parameters obtained using dynamic-contrast-enhanced MRI. The proposed approach may help in assisting radiologists, by defining precise tumor boundaries and hence, results in improving patients’ quality of life and overall survival.
Introduction
Gliomas originating from glial cells are the most commonly occurring primary brain tumors1-3. Low-grade gliomas (LGGs/grade-I & grade-II) in comparison to high-grade gliomas (HGGs/grade-III & grade-IV) have a relatively good prognosis and prolonged survival1,4. Diagnosis and grading of gliomas involve the combination of imaging, histopathology, and molecular information. Reported studies have suggested that maximum safe surgical resection helps in increasing overall survival1,2. Nearly all the primary brain tumors produce edema1,2,5,6. Safe surgical resection or precisely directed radiation requires distinguished tumor cells from peritumoral edema1,2,5. Tumor cells may comprise enhancing and non-enhancing components. Due to imaging technique limitations, clinicians find difficulty in defining the border between the non-enhancing tumor and edema components1-3,7-9. A previously reported study, using quantitative perfusion MRI, had reported a machine learning classifier for differentiating edema and non-enhancing tumor components of HGG2. For classifier development, edema was extracted from the patients with metastasis due to its clear demarcation with enhancing tumors. The current study aims to develop a machine learning-based classifier which uses quantitative perfusion parameters for differentiating non-enhancing tumor from edema in patients with LGGs cases.Methods
The current study included pre and post-surgery MRI data of 46 subjects (LGGs:23/metastasis:23). For classifier development, non-enhancing component of LGG and edema from metastasis patients were used. This is based upon the assumption that edema of metastasis and gliomas exhibits similar properties. For generating the non-enhancing tumor masks, post-surgery T1IR images were registered with corresponding pre-surgery T1-weighted images followed by skull stripping with the help of SPM (Statistical Parametric Mapping) software. The procedure for generating non-enhancing tumor masks for both the cases (enhancing and non-enhancing LGGs) was described in the presented flowchart (figure-1). For generating the edema masks, pre-surgery FLAIR and T1 contrast enhanced (T1CE) images were registered with the corresponding pre-surgery T1-weighted sequence and made skull-stripped. The procedure for generating edema masks was described in the presented flowchart (figure-1).The resulted segmented hypo-intense post-surgery T1IR and edema masks were further eroded to reduce the partial volume effect, hence resulting in nearly pure non-enhancing tumor and edema masks. These eroded non-enhancing tumor and edema masks were also verified by the radiologist and appropriate corrections were made, followed by quantitative perfusion parameters (QPP) computation with the help of inhouse developed perfusion-tool software. QPP includes hemodynamic (cerebral blood volume, cerebral blood flow), tracer kinetic (Ktrans,Ve, Vp , Kep) and other parameters (BAT, BETA, Slope-1 and Slope-2). Parameters computed for each pixel of every tumor mask, were arranged into ten column vectors. This process was repeated for every tumor case considered (dataset was named as DStmr). All the labels were put ‘1’ in the label field. Similar procedure was followed for edema mask hence resulting in dataset DSedm . Only difference was in the label field i.e., all labels were put ‘-1’ in the label field of DSedm. The dataset DStmr and DSedm were combined to form the dataset DSfnl followed by further division into training and testing set. Training set contains 40 cases (20 cases of each class) and testing set contains 6 cases (3 cases of each class). Based on the feature importance (computed with help of random-forest algorithm), sequential forward feature selection approach was implemented. The 10-fold cross-validation was performed on training dataset. Algorithms i.e., logistic regression (LR), k-nearest neighbor (KNN), random forest (RF) and linear SVM (kernel= ‘linear’), were considered for developing the machine learning based model.Results
Observed accuracy percentage in training, validation and testing set were 85.3, 80.24 and 83.6, respectively when model was trained using LR approach. The AUC value equals to 0.91 and 0.93 were recorded on training and test data (LR approach). The sensitivity and specificity values equal to 87.5, 81.6, 90.2 and 80.5 were recorded on training and testing set respectively. Table-II presents the summary of results recorded for considered different ML approaches. RF and Coarse KNN performed slightly poor on test dataset in comparison to LR and linear SVM approach. Figure-2 shows the LR based model performance (visual) on randomly considered test set images of each class against ground truth.Discussion
Differentiation between non-enhancing tumor and edema components is difficult on conventional MR images. In order to overcome this limitation, machine leaning based classifiers were developed in the current study. Generating tumor masks from post-surgery T1IR images can be thought as an alternative method of preparing tumor masks which can be considered more accurate in comparison to, what is defined manually by radiologists on pre-surgery images. For generating the edema mask, metastasis cases were considered. Use of Adaptive k-means clustering approach makes the mask segmentation procedure fast in comparison to manual mask segmentation procedure. It was noted that for the considered dataset, LR algorithm performs better. Proposed, approach should be evaluated on greater number of subjects, which might increase the accuracy.Conclusion
The presented study proposed a machine learning based approach for distinguishing non-enhancing tumor from edema in LGGs patients using quantitative perfusion parameters. The proposed approach may help in assisting radiologists, in order to define precise tumor region boundaries and hence resulting in improved patients’ quality of life and overall survival.Acknowledgements
Author acknowledge funding support from SERB-DST project nu CRG/2019/005032.
References
[1] D. A. Forst, B. V. Nahed, J. S. Loeffler, and T. T. Batchelor, “Low-Grade Gliomas,” Oncologist, vol. 19, no. 4, p. 403, Apr. 2014, doi: 10.1634/THEONCOLOGIST.2013-0345.
[2] A. Sengupta et al., “On differentiation between vasogenic edema and non-enhancing tumor in high-grade glioma patients using a support vector machine classifier based upon pre and post-surgery MRI images,” Eur. J. Radiol., vol. 106, no. 299, pp. 199–208, 2018, doi: 10.1016/j.ejrad.2018.07.018.
[3] M. Artzi, G. Liberman, D. T. Blumenthal, O. Aizenstein, F. Bokstein, and D. Ben Bashat, “Differentiation between vasogenic edema and infiltrative tumor in patients with high-grade gliomas using texture patch-based analysis,” J. Magn. Reson. Imaging, vol. 48, no. 3, pp. 729–736, 2018, doi: 10.1002/jmri.25939.
[4] Q. Yang, H. Zhang, J. Xia, and X. Zhang, “Evaluation of magnetic resonance image segmentation in brain low-grade gliomas using support vector machine and convolutional neural network,” Quant. Imaging Med. Surg., vol. 11, no. 1, pp. 300–316, 2021, doi: 10.21037/QIMS-20-783.
[5] K. I. Morita, H. Matsuzawa, Y. Fujii, R. Tanaka, I. L. Kwee, and T. Nakada, “Diffusion tensor analysis of peritumoral edema using lambda chart analysis indicative of the heterogeneity of the microstructure within edema,” J. Neurosurg., vol. 102, no. 2, pp. 336–341, 2005, doi: 10.3171/jns.2005.102.2.0336.
[6] H. J. Reulen, “Vasogenic brain oedema: New aspects in its formation, resolution and therapy,” Br. J. Anaesth., vol. 48, no. 8, pp. 741–752, 1976, doi: 10.1093/bja/48.8.741.
[7] A. L. and F. Gaillard, “Non-Contrast-Enhancing Tumor : A New Frontier in,” Am. J. Neuroradiol., pp. 1–8, 2019.
[8] M. I. Sharif, J. P. Li, M. A. Khan, and M. A. Saleem, “Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images,” Pattern Recognit. Lett., vol. 129, pp. 181–189, Jan. 2020, doi: 10.1016/j.patrec.2019.11.019.
[9] R. Zaouche et al., “Segmentation of low-grade gliomas in MRI: Phase based method,” 2nd Int. Conf. Adv. Technol. Signal Image Process. ATSIP 2016, pp. 97–102, 2016, doi: 10.1109/ATSIP.2016.7523061.
Figures
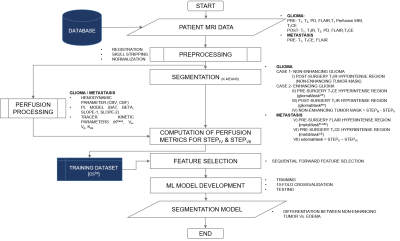
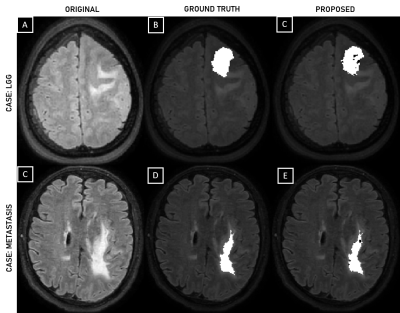
Figure 2. FLAIR images of LGG and metastasis cases (A, C), respectively; Ground truth i.e. non-enhancing tumor and edema, overlaid on FLAIR images of LGG and metastasis cases (B, D), respectively; Classifier predicted mask (LR approach) i.e. non-enhancing tumor and edema overlaid on FLAIR images of LGG and metastasis cases, respectively (C, E).