2723
Microscopically Anisotropic and Randomly Oriented 3D Printed MRI Phantom for Size, Orientation, and Anisotropy Validation in Diffusion Imaging
Velencia Witherspoon1, Michal Komlosh1,2,3, Dan Benjamini1,2,3, Peter Basser1,3, and Nick Lavrik4
1Section on Quantitative Imaging and Tissue Sciences, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, United States, 2Center for Neuroscience and Regenerative Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD, United States, 3The Henry M. Jackson Foundation for the Advancement of Military Medicine (HJF), Bethesda, MD, United States, 4The Center for Nanophase Materials Sciences (CNMS) at Oak Ridge National Laboratory (ORNL), Oakridge, TN, United States
1Section on Quantitative Imaging and Tissue Sciences, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, United States, 2Center for Neuroscience and Regenerative Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD, United States, 3The Henry M. Jackson Foundation for the Advancement of Military Medicine (HJF), Bethesda, MD, United States, 4The Center for Nanophase Materials Sciences (CNMS) at Oak Ridge National Laboratory (ORNL), Oakridge, TN, United States
Synopsis
Microscopically anisotropic but globally isotropic DTI MRI phantom was designed and fabricated using a 2 Photon Nanoscribe 3D Printer. The capillary array blocks were measured aligned and randomly oriented to determine the ability of ddPFG and DTI to characterize local anisotropy in a globally isotropic system. These experiments combined computational methods can provide MRI phantoms that can be employed to validate novel or proposed microstructure imaging experiments.
Introduction
Diffusion imaging methods are commonly employed to infer changes in tissue microstructural features due to pathology. Quantities like the mean apparent diffusion coefficient (mADC), and the fractional anisotropy (FA) inter alia are inherently influenced by tissue morphology such as effective pore size, shape, and connectivity[2,3,5]. As diffusion-related parameters are increasingly being applied to provide additional image contrast, where they are used as pathological biomarkers, and indicators of tissue microstructure, it is vital to quantitatively determine the performance of these methods on model phantom/material systems that mimic salient features of these tissue.[1] There is a current drive in the methods development community to robustly determine and differentiate pore size distributions, combine features of microscopic anisotropy with random orientation [3,4,5], and assess the effect of diffusion exchange [6] to the sensitivity of diffusion methods. However, there is a tendency to transition directly from theory to tissue-based applications without taking the necessary intermediate step to vet such methods. To achieve a better quantitation in single subject, longitudinal and particularly multi-site studies, it is important to standardize MR scanner data and the pipelines used to process them [1,7]. A key element in this is the development of well-characterized MRI phantoms that exhibit salient characteristics of target tissues. Several notable MRI phantoms have been fabricated to represent microscopically anisotropic but macroscopically isotropic grey matter tissue [8-16,19]. For example, the glass capillary array phantom developed by Komlosh et al. [13] was used to vet pore size, pore size distribution, and fractional anisotropy determination via double pulsed-field gradient (dPFG) MR methods. These phantoms, although functional, often suffer from susceptibility contrast between phantom material and water when compared to biological tissues [8,17], material robustness [14], safety issues, and transportability [12]. To address this, we have utilized advanced 3D printing technology with +/-100 nm pore shape accuracy to produce MRI phantoms that possess microscopic anisotropy as are an increasing number of tissues, including in the brain. Complex tissues are often composed of morphological features that vary in their microscopic FA (μFA), compartment size distributions, and orientations. This method can greatly advance the diagnostic capabilities of MRI to differentiate axon size and shape distributions in white matter and randomly oriented microscopically anisotropic structure, such as what is observed in gray matter [19]. In order to advance the application of diffusion imaging methods as tissue biomarkers, we must also advance the complexity of our phantoms used to validate them.Methods
We printed MRI Phantoms using the 2-Photon Lithography Nanoscribe at The Center for Nanophase Materials Sciences (CNMS) at Oak Ridge National Laboratory (ORNL) to 3D print aligned capillary arrays, of 10 μm inner diameter (ID), in blocks of 300 μm x 300 μm x 300 μm. Scanning electron microscopy (SEM) images were taken before and after the MRI experiments. The blocks were plasma treated to make the surface hydrophilic and then loaded with water in a glass tube. The aligned capillary pore size was measured using a dPFG spin-echo imaging sequence, using a previously proposed protocol [20]. These same blocks were then removed from their support, randomly distributed in a glass tube, as in an emulsion, and re-measured using a dPFG and a standard diffusion tensor imaging (DTI) sequence. All MRI measurements were performed on the Avance III 7T Bruker scanner equipped with a Micro 2.5 microimaging gradient set. A mixed model of restricted and Gaussian diffusion [21] was used to fit the dPFG data, with the restricted diffusion modeled to arise from a cylindrical pore geometry [22]. We utilized custom MATLAB code to estimate the voxel-wise average pore size accounting for the diffusion gradients profile. We used TORTOISE to produce DTI images.Results
The 3D printed method results in homogenous pore size, with mean and standard deviation, is described by the nominal value of 10.7 +/- 0.323 μm obtained from microscopy images (Figure 1A), as well as a single layer or well-aligned pores used for the dPFG acquired ROI (Figure 1B). The voxelwise pore size estimation resulted in fairly homogenous pore size values, with a mean and standard deviation of 8.33 +/- 1.45 μm (Fig. 1C).In Figure 2, Diffusion-Encoded-Color (DEC) (A) map is juxtaposed with the FA map (B) at 50 μm resolution determined by the 3D-DTI. This validates that the blocks are randomly oriented and microscopically anisotropic, supported by the SEM images (A, B) in Figure 1. When the entire slice is used as an ROI from the DTI image the AD is 1.10 x10-9 m2/s, the RD is 9.81 x10-9 m2/s and the FA is 0.070, showing macroscopic isotropy.
Conclusion
Microscopically anisotropic but macroscopically isotropic DTI MRI phantom was designed and fabricated using a 2 Photon Nanoscribe 3D Printer. The capillary array blocks were measured aligned and randomly oriented to determine the ability of dPFG and DTI to characterize local anisotropy in a globally isotropic system. Although these blocks were printed with 10 μm pore diameter, it is possible to reduce or increase this diameter to better reflect neurological structural features found in the Central and Peripheral Nervous Systems. These experiments combined computational methods can provide MRI phantoms that can be employed to validate novel or proposed microstructure imaging experiments.Acknowledgements
This work was supported by the Intramural Research Program (IRP) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, “Connectome 2.0: Developing the next generation human MRI scanner for bridging studies of the micro-, meso- and macro-connectome”, NIH BRAIN Initiative 1U01EB026996-01, and the CNRM Neuroradiology/Neuropathology Correlation/Integration Core, 309698-4.01-65310, (CNRM-89-9921).A portion of this research was conducted at the Center for Nanophase Materials Sciences, which is a DOE Office of Science User FacilityReferences
- Souza EM, Costa ET, Castellano G. “Phantoms for diffusion-weighted imaging and diffusion tensor imaging quality control: a review and new perspectives.” Res Biomed Eng. 2017; 33(2):156-165.
- Szczepankiewicz, Filip et al. “Quantification of microscopic diffusion anisotropy disentangles effects of orientation dispersion from microstructure: applications in healthy volunteers and in brain tumors.” NeuroImage vol. 104 (2015): 241-52.
- Magdoom, K.N., Pajevic, S., Dario, G. et al. A new framework for MR diffusion tensor distribution. Sci Rep 11, 2766 (2021).
- Martin J., Endt S., Wetscherek A. et. al. , Contrast-to-noise ratio analysis of microscopic diffusion anisotropy indices in q-space trajectory imaging Z. Med. Phys., 30 (2020), pp. 4-16
- Ianuş A., Drobnjak I., Alexander D.C. (2015) Model-Based Estimation of Microscopic Anisotropy in Macroscopically Isotropic Substrates Using Diffusion MRI. In: Ourselin S., Alexander D., Westin CF., Cardoso M. (eds) Information Processing in Medical Imaging. IPMI 2015. Lecture Notes in Computer Science, vol 9123. Springer, Cham.
- Bai R., Li Z., Sun C., et al. Feasibility of filter-exchange imaging (FEXI) in measuring different exchange processes in human brain, NeuroImage, Volume 219, 2020
- Zhu T., Hu R., Qiu X. et al. ,"Quantification of Accuracy and Precision of Multi-Center DTI Measurements: A Diffusion Phantom and Human Brain Study," Neuroimage, vol. 56, no. 3, p. 1398–1411, 2011.
- Farrher E, Lindemeyer J, Grinberg F, Concerning the matching of magnetic susceptibility differences for the compensation of background gradients in anisotropic diffusion fibre phantoms. (2017) PLoS ONE 12(5): e0176192.
- Gatidis S, Schmidt H, Martirosian P, et al. Development of an MRI phantom for diffusion-weighted imaging with independent adjustment of apparent diffusion coefficient values and T2 relaxation times. Magnetic Resonance in Medicine. 2014; 72(2):459-63.
- Kłodowski K, Krzyżak AT. Innovative anisotropic phantoms for calibration of diffusion tensor imaging sequences. Magn Reson Imaging. 2016 May;34(4):404-9.
- Suzuki, M., Moriya, S., Hata, J. et al. Development of anisotropic phantoms using wood and fiber materials for diffusion tensor imaging and diffusion kurtosis imaging. Magn Reson Mater Phy 32, 539–547 (2019). https://doi.org/10.1007/s10334-019-00761-3
- Komlosh, ME, Horkay, F, Freidlin R, et. al., Detection of microscopic anisotropy in gray matter and in a novel tissue phantom using double Pulsed Gradient Spin Echo MR, (2007) J Magn Reson 189 7 38-45
- Komlosh ME, Özarslan E, Lizak MJ, et al. Pore diameter mapping using double pulsed-field gradient MRI and its validation using a novel glass capillary array phantom. J Magn Reson. 208(1) (2011):128-135.
- Lätt, Jimmy, Nilsson, Markus, Rydhög, Anna et al.(2007). Effects of restricted diffusion in a biological phantom: A q-space diffusion MRI study of asparagus stems at a 3T clinical scanner. Magma (New York, N.Y.). 20. 213-22.
- Nilsson, M., Larsson, J., Lundberg, D., et. al. (2018). Liquid crystal phantom for validation of microscopic diffusion anisotropy measurements on clinical MRI systems. Magnetic resonance in medicine, 79(3), 1817-1828
- Qin, E. C., Sinkus, R. , Geng, G. , et. al (2013), Combining MR elastography and diffusion tensor imaging for the assessment of anisotropic mechanical properties: A phantom study. J. Magn. Reson. Imaging, 37: 217-226
- Laun FB, Huff S, Stieltjes B. On the effects of dephasing due to local gradients in diffusion tensor imaging experiments: relevance for diffusion tensor imaging fiber phantoms. Magn Reson Imaging. 2009 May;27(4):541-8.
- Westin, C. F., Knutsson, H., Pasternak, O., Szczepankiewicz, et. al.(2016). Q-space trajectory imaging for multidimensional diffusion MRI of the human brain. NeuroImage, 135, 345-362.
- Komlosh ME, Benjamini D, Barnett AS, et. al. Anisotropic phantom to calibrate high-q diffusion MRI methods. J Magn Reson. 275 (2017):19-28
- Benjamini D, Komlosh ME, Basser PJ, et al. Nonparametric pore size distribution using d-PFG: comparison to s-PFG and migration to MRI. J Magn Reson. 246 (2014):36-45.
- Assaf Y, Basser PJ. Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. Neuroimage. 27(1) (2005) : 48-58.
- Grebenkov, Denis S. “Laplacian Eigenfunctions in NMR. I. A Numerical Tool.” Concepts in Magnetic Resonance Part A 32 (2008): 277-301
Figures
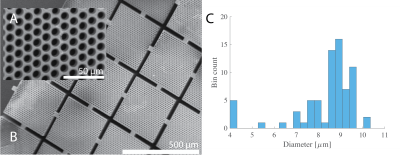
Figure 1 (A) Inset of scanning electron microscopy (SEM) images of the 3D printed capillary array (B). (C) Resulting pore diameter histogram calculated from dPFG experiments with axial slices.
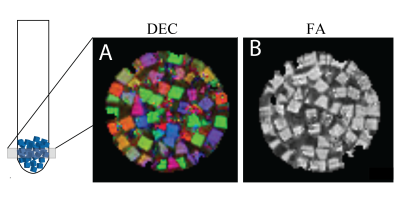
Figure 2 Diffusion-Encoded-Color (DEC) map (A) and fractional anisotropy (FA) map (B) determined by 3D-DTI-MAP MRI with diffusion-weighted EPI acquisition
DOI: https://doi.org/10.58530/2022/2723