2716
FAR-CEST: Fast Acquisition and Reconstruction for Chemical Exchange Saturation Transfer (CEST) imaging using a Deep-Learning Approach1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China, 2Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 3Xi’an Key Lab of Radiomics and Intelligent Perception, School of Information Sciences and Technology, Northwest University, Xi'an, China
Synopsis
We developed a Fast Acquisition and Reconstruction CEST (FAR-CEST) method at 3T human scanners, based on a deep learning approach. A 10X accelerated acquisition was achieved, which under-sampled K-space using a randomized Cartesian pattern of variable density. To fully utilize the correlation among saturation offset dimension, especially to compensate for sparsely-sampled K-space edge, a 3D-Res-Unet model was trained for reconstruction. Results on healthy adult brain suggested that FAR-CEST can produce high quality saturation-weighted images and Z-spectra,but the CEST contrast slightly altered. The highly-acceleration feature of FAR-CEST has been initially validated, yet still require improvement on reconstruction accuracy.
Introduction
Chemical exchange saturation transfer (CEST) MRI is an emerging ‘label-free’ molecular imaging tool available in clinic. Usually CEST requires collection of several saturation weighted images, each with a seconds-long saturation preparation for signal amplification. Therefore, current acquisition protocols face obstacles of long scan time, low-resolution images and limited number of saturation frequencies[1-3]. Herein, we utilized randomized K-space under-sampling and the redundancy among the multiple saturation-weighted images, to achieve Fast Acquisition and Reconstruction CEST imaging (FAR-CEST) through a deep-learning method.Methods
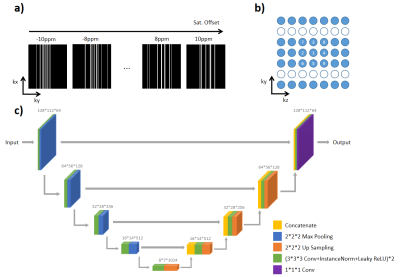
Under-sampling pattern of K-spaceWith the sparse feature of CEST MRI signal[4], a high acceleration factor could be achieved by fully utilizing the correlation of CEST images along the offset dimension (Figure 1a). For each slice pre-labeled by different saturation offsets, a randomized cartesian under-sampling pattern is designed, with sampling density following a Poisson distribution at K-space center. For K-space edges which had low sampling density, the randomness among images with the saturation offsets will help CNN reconstruct the missed line better. Besides, to maximize SNR of image, a low-high order was applied in acquisition (Figure 1b).
Data Acquisition
Twelve healthy volunteers were scanned on a 3T scanner (Ingenia CX 3.0T; Philips Medical Systems, Best, The Netherlands), using a body coil for transmission and a 32-channel head coil for reception. Among all subjects, 10 were acquired using fully-sampled CEST sequence that took 15 mins each; For two of the subjects, apart from above -mentioned fully-sampled data, prospective 10X under-sampled acquisition was performed, only costing1 min 53 s. All subjects signed the written informed consent. The CEST sequence used a saturation duration of 2 seconds and power of 1 uT, followed by TSE readouts. In total 32 saturation offsets were collected for Z-spectra fitting, distributed from -10 ppm to 10ppm. Other parameters are: FOV=230×201×60 mm3, spatial resolution=1.8×1.8×6 mm3, slice number=10, TR/TE=5.5 s/7.8 ms, TSE factor=174.
Data Preprocessing
To generate the input data for network training, all full-sampled k-space data were retrospectively under-sampled and reconstructed with inverse Fast Fourier transform according to Equation 1.
$$y=\sqrt{\sum_{i}^{} \left|F^{H}Px_{i}\right|^{2}} $$where xi is full sampled K-space data of ith channel, P is an under-sampling mask, and FH means Inverse Fast Fourier transform (IFFT) from K-space to image space. All the images were under-sampled by 10 times(10X) and the network input were CEST data for each slice, with a size of Nx X Ny X Nw(Nw: Number of Saturation offsets)
Network Training
A 3D Res-U-Net network[5] was implemented for reconstruction task (Figure 1c), in which all convolution operations of U-Net were replaced by residual blocks. Considering each image slice as a training datum, the inputs of network are 3D patches with size of 128*112*32, to simultaneously learn the spatial and frequency features, and their correlations. Each subject contained 9 slices, the number of training data was 10 subjects, and the number of test data was 2 subjects. Learning rate was set to 1e-4, then decayed by 10 times at 30 and 150 epochs. The number of training epochs was set as 300. The employed loss function was mean squared error (MSE), and the employed optimizer was Adam.
Results and Discussion
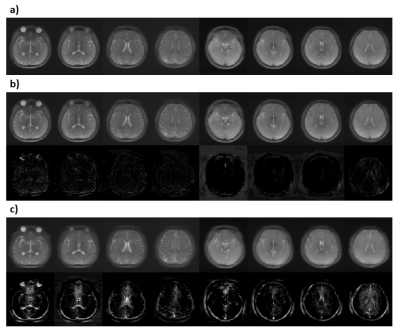
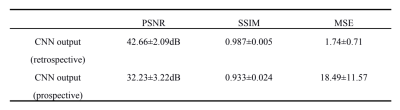
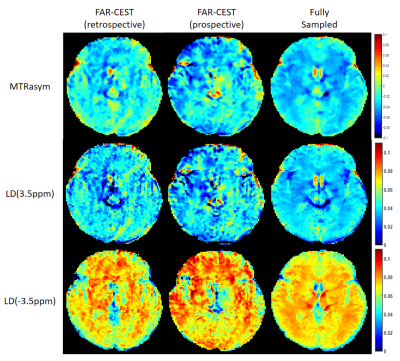
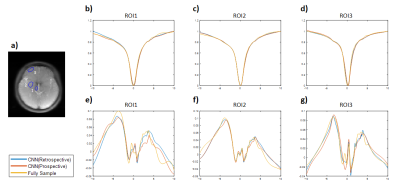
We evaluated the performance of FAR-CEST, with regular Fourier-transform based reconstruction from fully-sampled data as the golden standard. Figure 2 compared the saturation-weighted images at 3.5 ppm obtained from the full-sampled data, the FAR-CEST reconstruction from the retrospective data and from the prospective data, respectively. The difference between FAR-CEST images and fully-sampled golden standard suggested the good recovery of anatomy structure , but the prospective images displayed clear signal difference at ventricles, eye balls and brain boundaries . From Table 1, the CNN with prospective under-sampling achieved 32.23 dB of PSNR, 0.933 of SSIM, and 18.49 of MSE, which is closed to the golden standard. We further calculated the contrast maps through the widely-used quantification methods, i.e., MTR asymmetry and Lorentzian difference (LD) (Figure 3), which indicted retrospective and prospective results were consistent with fully-sampled results but were noisier and displayed slightly altered contrast from the golden standard. Despite averaged Z-spectra of ROIs from FAR-CEST images were very close to the golden standard, the quantitative performance of FAR-CEST still need improvement, since CEST observed signal changes of a few percent.In this initial try, for proving the concept, only 3D-Res-U-net reconstruction was employed for a 10X acceleration, resulting in quantitative images slightly altered from the golden standard. In future optimization of the network type and architecture, as well as the growing size of training data, could improve the FAR-CEST performance. The constrains of Z-spectral line-shape could also been added to loss function, which may lead the network for more attentions on spectra reconstruction. Also noted that in the current implementation, the under-sampling was actually performed on kx, ky and offsets dimension, excluding kz dimension. Moreover, our approach could be easily combined with other CEST acceleration methods to improve the quantification performance, such as sequence-based methods and compressed sensing-based methods.
Conclusion
Herein, we initially validated the fast acquisition and reconstruction for CEST (FAR-CEST) Imaging using a deep-learning approach. The results suggested that FAR-CEST could achieve 10X acceleration, producing similar quality images to the fully-sampled golden standard, which may help clinical promotion of CEST imaging.Acknowledgements
The authors acknowledged funding from National Natural Science Foundation of China (82071914). and the startup package from Tsinghua University to Dr. Song.References
[1] Y. Zhang, H.-Y. Heo, D.-H. Lee, S. Jiang, X. Zhao, P. A. Bottomley, et al., "Chemical Exchange Saturation Transfer (CEST) Imaging With Fast Variably-accelerated Sensitivity Encoding (vSENSE)," Magnetic Resonance in Medicine, vol. 77, pp. 2225-2238, Jun 2017.
[2] C. Guo, J. Wu, J. T. Rosenberg, T. Roussel, S. Cai, and C. Cai, "Fast chemical exchange saturation transfer imaging based on PROPELLER acquisition and deep neural network reconstruction," Magnetic Resonance in Medicine, vol. 84, pp. 3192-3205, Dec 2020.
[3] H. She, J. S. Greer, S. Zhang, B. Li, J. Keupp, A. J. Madhuranthakam, et al., "Accelerating chemical exchange saturation transfer MRI with parallel blind compressed sensing," Magnetic Resonance in Medicine, vol. 81, pp. 504-513, Jan 2019.
[4] M. Lustig, D. Donoho, and J. M. Pauly, "Sparse MRI: The application of compressed sensing for rapid MR imaging," Magnetic Resonance in Medicine, vol. 58, pp. 1182-1195, Dec 2007.
[5] X. Xiao, S. Lian, Z. Luo, S. Li, and Ieee, "Weighted Res-UNet for High-quality Retina Vessel Segmentation," in 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, PEOPLES R CHINA, 2018, pp. 327-331.
Figures