2713
Acceleration of Quantitative Semisolid MT/CEST Imaging using a Generative Adversarial Network (GAN-CEST)1Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA, United States, 2Department of Neuroradiology, Friedrich-Alexander Universität Erlangen-Nürnberg (FAU), University Hospital Erlangen, Erlangen, Germany, 3Magnetic Resonance Center, Max Planck Institute for Biological Cybernetics, Tubingen, Germany, 4Department of Biomedical Magnetic Resonance, University of Tübingen, Tübingen, Germany, 5Cardiovascular Research Center, Cardiology Division, Massachusetts General Hospital, Charlestown, MA, United States, 6Health Science Technology, Harvard-MIT, Cambridge, MA, United States
Synopsis
Quantitative metabolite concentration and pH biomarker maps, as provided by semisolid MT/CEST-MR-Fingerprinting (MRF), constitute a useful means for determining the molecular origin of pathology. However, the lengthy dictionary generation time and the prolonged 3D acquisition time may hinder clinical dissemination. Here, we developed a generative adversarial network (GAN), aimed to drastically shorten the 3D semisolid MT/CEST-MRF acquisition time and circumvent the need for dictionary generation. In-vitro and in-vivo experiments in 4 volunteers and a patient were conducted at 3 different sites using 3 different scanner models, showing substantial reduction in scan time, while retaining a good agreement with ground-truth reference.
Introduction
Magnetic resonance fingerprinting (MRF) is a powerful imaging strategy, capable of rapidly extracting quantitative tissue parameter maps1. MRF was recently expanded for the quantification of additional tissue parameters, including the chemical exchange properties of the semisolid-MT and mobile proteins, lipids, and metabolites, detectable via the chemical exchange saturation transfer (CEST) mechanism2,3. Such molecular imaging scenarios, which involve multiple proton pools, impose additional challenges on the implementation of MRF. In particular, the large number of tissue parameters exponentially increases the size of the simulated dictionary, which in turn increases the parameter matching time or the time for training a reconstruction neural-network. In addition, CEST MRF requires sufficient amplification of the signals stemming from the low millimolar concentration of metabolites/proteins, which requires saturation pulses of more than a second-long, and hence, a longer acquisition time compared to water-pool MRF4.Recently, deep learning-based methods for reconstruction of semisolid MT/CEST-MRF data were developed5,6, to shorten the parameter quantification time. Although these strategies have demonstrated promising results, the 3D acquisition time was still considerably long, limiting the integration in clinical routine.
The purpose of this work is to allow for substantially shorter semisolid-MT/CEST-MRF acquisition, by reducing the number of raw images used to encode the quantitative molecular information. We employed a generative adversarial network supervised framework (GAN-CEST), which learns the mapping from a reduced input data space to the quantitative exchange parameter space.
Methods
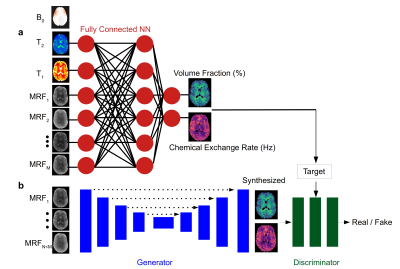
GAN-CEST ArchitectureA supervised learning framework (Fig. 1) was designed based on the conditional GAN architecture7. The generator was a U-Net convolutional network aiming to synthesize proton exchange parameter maps, for either the semisolid MT or the CEST compound exchangeable protons, for a given partial subset of N raw MRF encoding images. The discriminator aimed to predict whether the images are the ‘real’ corresponding quantitative images, or a ‘fake’ (generator synthesized maps). The ground truth was obtained by a dictionary-trained fully connected CEST-MRF neural-network5 that received the full M-length acquisition schedule as input (Fig. 1a).
Phantom Imaging
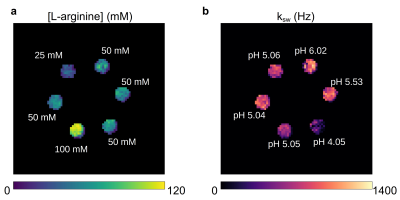
For obtaining a preliminary yet well-controlled validation of the quantitative output obtained by GAN-CEST, a set of phantoms was assembled with 25-100 mM L-arginine concentrations, at pH 4-6. Imaging was performed at two different sites, using a 3T Siemens Prisma or Skyra scanner as described in the next section, with the number of raw images set to N=M=30 iterations.
Human Imaging
All scans were performed with IRB approval and informed consent, using a 3D whole-brain protocol (1.8 mm isotropic resolution), with a single-shot EPI readout, implemented using the pulseq-CEST framework8,9. For system’s training, 4 healthy volunteers were scanned using a 3T Siemens Prisma or Skyra scanners at two imaging sites, creating a total of 493 training images. For creating a challenging validation setting, a GBM patient was scanned at a third site, using a 3T Siemens Trio scanner, creating a total of 85 validation images. An MRF acquisition schedule of M=30 iterations, with various saturation pulse powers and frequency offsets was employed, aiming to quantify the semisolid-MT exchange parameters.
Data processing
All images were motion-corrected and registered using Elastix. Gray-matter and white-matter segmentation was performed using SPM. Quantitative reference CEST-MRF maps were obtained using a fully connected neural network5, trained on simulated dictionaries while considering all raw input measurements and receiving T1, T2, and B0 values as direct input (for further improving the accuracy of the human data ground truth reference, Fig. 1a). N=20 raw images were used for GAN-CEST in-vivo quantification.
Results
In-vitro studyA representative validation image-set for the GAN-CEST-based exchange parameter quantification in the phantom experiment is shown in Fig. 2. A good agreement was obtained between the estimated L-arginine concentrations and the known concentrations (Pearson’s r = 0.992), and between the estimated proton exchange rates and the reference QUESP calculated values (Pearson’s r = 0.622).
Human study
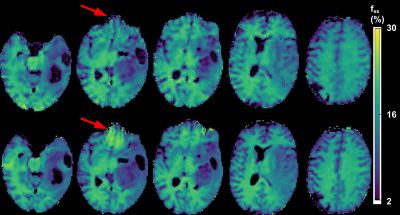
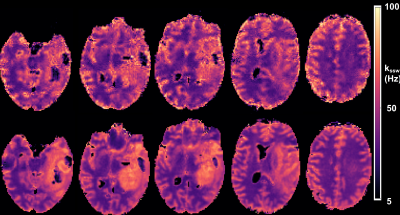
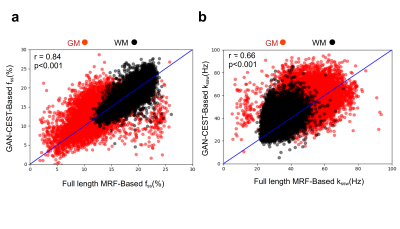
A comparison between the GAN-CEST results and the full-length MRF-based reference for five representative slices is available in Fig. 3 (fss) and Fig. 4 (kssw). Surprisingly, even-though the training cohort included only healthy volunteers, GAN-CEST was able to output visually similar maps, even in complex tumor and edema containing image slices. Moreover, the method was able to mitigate B0 inhomogeneity effects, without receiving explicit B0 information (red arrows in Fig. 3). The absolute error in estimating the semisolid proton volume fraction (fss) and exchange rate (kssw), for the entire brain was 1.97±2.28 (%) and 9.69±10.38 Hz, respectively. The resulting WM/GM fss estimated by the GAN-CEST approach for all non-tumor-containing slices was 18.12±2.57 / 13.03±2.86 %, compared to 18.73±2.01 / 12.36±2.72 % using the full-length MRF. The WM/GM kssw estimated by the GAN-CEST approach was 36.63±8.2 / 52.32±10.57 Hz, compared to 33.88±5.23 / 49.06±8.54 Hz, by the full-length MRF reference, with significant correlation between individual pixels values obtained by both methods (Fig. 5). The acquisition schedule used for GAN-CEST was 33% shorter than the full-length MRF schedule and did not require the separate acquisition of T1/T2/B0 maps (as opposed to CEST-MRF, Fig. 1a), further reducing the total scan time.
Conclusion
GAN-CEST MRF can substantially reduce the acquisition time of quantitative semisolid MT/CEST mapping, while retaining the performance even for unseen pathologies and different sites and scanner models.Acknowledgements
National Institutes of Health Grant/Award Numbers: R01CA203873, P41-RR14075. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 836752 (OncoViroMRI). This paper reflects only the author’s view, and the European Research Executive Agency is not responsible for any use that may be made of the information it contains.References
1. Ma, D., Gulani, V., Seiberlich, N., Liu, K., Sunshine, J.L., Duerk, J.L. and Griswold, M.A., 2013. Magnetic resonance fingerprinting. Nature, 495(7440), pp.187-192.
2. Cohen, O., Huang, S., McMahon, M.T., Rosen, M.S. and Farrar, C.T., 2018. Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF). Magnetic Resonance in Medicine, 80(6), pp.2449-2463.
3. Heo, H.Y., Han, Z., Jiang, S., Schär, M., van Zijl, P.C. and Zhou, J., 2019. Quantifying amide proton exchange rate and concentration in chemical exchange saturation transfer imaging of the human brain. Neuroimage, 189, pp.202-213.
4. Perlman, O., Herz, K., Zaiss, M., Cohen, O., Rosen, M.S. and Farrar, C.T., 2020. CEST MR‐Fingerprinting: Practical considerations and insights for acquisition schedule design and improved reconstruction. Magnetic Resonance in Medicine, 83(2), pp.462-478.
5. Perlman Or, Ito Hirotaka, Herz Kai, et al. Quantitative imaging of apoptosis following oncolytic virotherapy by magnetic resonance fingerprinting aided by deep learning Nature Biomedical Engineering. https://doi.org/10.1038/s41551-021-00809-7. 25 2021;
6. Kim, B., Schär, M., Park, H. and Heo, H.Y., 2020. A deep learning approach for magnetization transfer contrast MR fingerprinting and chemical exchange saturation transfer imaging. Neuroimage, 221, p.117165.
7. Isola, P., Zhu, J.Y., Zhou, T. and Efros, A.A., 2017. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 1125-1134).
8. Herz, K., Mueller, S., Perlman, O., Zaitsev, M., Knutsson, L., Sun, P.Z., Zhou, J., van Zijl, P., Heinecke, K., Schuenke, P. and Farrar, C.T., 2021. Pulseq‐CEST: Towards multi‐site multi‐vendor compatibility and reproducibility of CEST experiments using an open‐source sequence standard. Magnetic Resonance in Medicine.
9. Zaiss, M., Ehses, P. and Scheffler, K., 2018. Snapshot‐CEST: optimizing spiral‐centric‐reordered gradient echo acquisition for fast and robust 3D CEST MRI at 9.4 T. NMR in Biomedicine, 31(4), p.e3879.
Figures