2691
Open Science Initiative for Perfusion Imaging (OSIPI): A community-led, open-source code library for analysis of DCE/DSC-MRI1Department of Radiation Oncology, the Netherlands Cancer Institute, Amsterdam, Netherlands, 2Neuroimaging Research, Barrow Neurological Institute, Phoenix, AZ, United States, 3Quantitative Biomedical Imaging Laboratory, Division of Cancer Sciences, University of Manchester, Manchester, United Kingdom, 4Radiology, Mayo Clinic, Rochester, MN, United States, 5Edinburgh Imaging and Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, United Kingdom, 6Department of Radiology and Nuclear Medicine, Amsterdam UMC, Amsterdam, Netherlands, 7Institute of Medical Physics, The University of Sydney, Sydney, Australia, 8Department of Oncology, Aarhus University, Aarhus, Denmark, 9Institute of Clinical Sciences, Department of Radiation Sciences, The University of Gothenburg, Gothenburg, Sweden, 10Manchester Academic Health Science Centre, Division of Informatics, Imaging, and Data Science, School of Health Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester, United Kingdom, 11Institute of Radiology, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 12Clinical Imaging, Genentech, Inc, South San Francisco, CA, United States, 13Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, United Kingdom, 14Edinburgh Imaging and Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, United Kingdom
Synopsis
A lack of validated, open-source code reduces the reliability of perfusion MRI, resulting in duplicate development. To address this problem, the Open Science Initiative for Perfusion Imaging (OSIPI) established a taskforce to collect, validate and harmonise such code. To date, 74 code contributions have been collected, with 14 of these tested. Source code and tests are published in an open-access repository. The OSIPI DCE/DSC-MRI code collection constitutes a valuable resource for researchers, and will ultimately be developed into a standardised, community-driven open-source code library.
Introduction
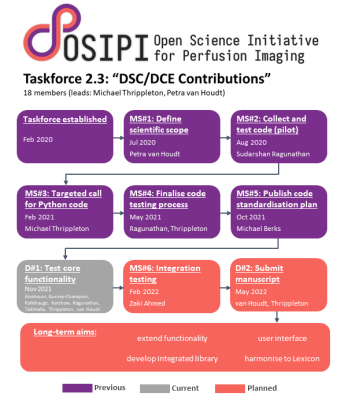
Perfusion MRI results show substantial variability dependent on the processing software used [1,2]. Furthermore, researchers expend substantial effort in writing bespoke code for their analyses because existing solutions are unavailable, unvalidated, difficult to use or provide insufficient functionality. Taskforce 2.3 of the Open Science Initiative for Perfusion Imaging (OSIPI) was therefore established to collect, validate and harmonise open-source code snippets for processing dynamic contrast-enhanced (DCE-) and dynamic susceptibility contrast (DSC-) MRI, as a resource for researchers and developers (Figure 1). Here we report current progress towards these ongoing objectives and present initial results.Methods
A public call for code was made via the ISMRM Perfusion Study Group in February 2021, and contributions were published in a public repository [3]. To provide greatest utility for the most users, code to perform the most common steps of DCE- and DSC-MRI processing pipelines was prioritised. Initial code collection was restricted to the Python language, since it is open-source, widely used and has many high quality scientific computing libraries.To validate code contributions, automated scientific testing was implemented within the online repository using the Pytest package and the Github Actions feature. The aim is to verify that code outputs match previously defined or measured reference values to within an acceptable tolerance defined by the taskforce, and to document the outputs of each contribution. Test data was taken from digital reference objects (DROs) and clinical scans. Reference values for in-vivo test data were obtained using separate, trusted software; for DRO data, reference values supplied with the DRO were used. Code for variable flip angle T1 mapping was validated using a Quantitative Imaging Biomarker Alliance DRO (version 3 [4]), and in-vivo brain [5] and prostate [6] scans. Code contributions implementing the extended Tofts pharmacokinetic model were validated using data from an anthropomorphic DRO [7].
The final objective is to harmonise and integrate code contributions into a coherent code library. During the present two-year cycle of the taskforce, we aimed to develop its outline structure.
Results
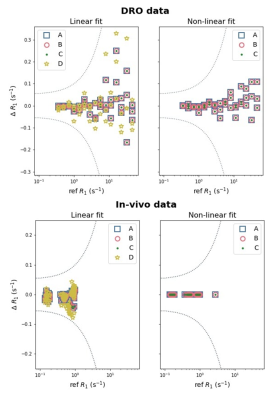
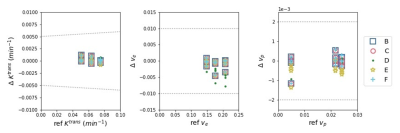
As of 1 November 2021, the repository hosts 74 distinct units of perfusion functionality contributed by 11 individuals and groups. This includes code implementing all core aspects of DCE- and DSC-MRI processing, for example T1 mapping, concentration estimation and perfusion parameter derivation, as shown in Table 1. Details of all collected code can be viewed in the online repository [3].Validation is in-progress across all core areas of functionality (Table 1). Preliminary results for seven T1 mapping implementations are shown in Figure 2. All results matched the reference values to within the tolerance, and multiple implementations of equivalent algorithms generated similar outputs. For the extended Tofts pharmacokinetic model, five different code contributions were validated. The resulting estimates of KTrans,ve and vp were within the tolerances (Figure 3).
As a step towards code harmonisation, a repository was established to host an integrated code library and an outline design for the library was proposed [8]. Briefly, community code contributions will be integrated into the OSIPI Perfusion Analysis Library, which will facilitate basic and advanced image processing functionality. The library will include low-level methods, processing pipelines and user interfaces; classes, methods and variables will map to concepts defined in the consensus-driven OSIPI DCE/DSC-MRI perfusion lexicon (Taskforce 4.2).
Discussion
DSC-MRI functionality is currently underrepresented in the repository; we therefore encourage new code contributions in this area. We also aim to collect code implementing additional aspects of perfusion processing, such as model selection, multiple pulse sequences and water exchange effects. Where Python code is unavailable, the taskforce may decide to translate or provide wrappers to contributions written in other languages such as Matlab and C++.Contributions tested thus far yielded results in good agreement with reference values. Testing is currently being expanded to cover broader parameter ranges, different pharmacokinetic models and other processing steps. A challenge of the pass-fail testing approach is that tolerances are subjective and difficult to determine, and binary test outputs provide limited useful information to potential users. We are therefore exploring complementary approaches for automated documentation and visualisation of code outputs.
Finally, the taskforce aims to enhance the value of the repository by integrating contributions into the OSIPI Perfusion Analysis Library, which will be maintained by the perfusion community to facilitate standardised processing and reporting of perfusion MRI.
Conclusion
The OSIPI Taskforce 2.3 code collection constitutes a valuable and growing resource to benefit researchers using contrast-based perfusion imaging. Our approach, which is based on open-source tools and free web-based services, may be an appropriate model for open science initiatives in other MRI subfields.Acknowledgements
We acknowledge the OSIPI membership and leadership for their suggestions and feedback, Ole Gunnar Johansen (University of Oslo) and Matthew Orton (Royal Marsden NHS Foundation Trust) for contributing additional code, and Joanna M. Wardlaw (University of Edinburgh) for providing test data.References
1. Beuzit et al., Dynamic contrast-enhanced MRI: Study of inter-software accuracy and reproducibility using simulated and clinical data. JMRI 2016; 43:1288.
2. Kudo et al., Accuracy and Reliability Assessment of CT and MR Perfusion Analysis Software Using a Digital Phantom. Radiology 2013; 267:201
3. https://github.com/OSIPI/DCE-DSC-MRI_CodeCollection/wiki
4. Daniel Barboriak, https://qidw.rsna.org/#collection/594810551cac0a4ec8ffe574
5. Clancy et al., Rationale and design of a longitudinal study of cerebral small vessel diseases, clinical and imaging outcomes in patients presenting with mild ischaemic stroke: Mild Stroke Study 3. Eur Stroke J 2020; 6:81.
6. Klawer et al., Improved repeatability of dynamic contrast-enhanced MRI using the complex MRI signal to derive arterial input functions: a test-retest study in prostate cancer patients. MRM 2019; 81:3358
7. Bosca and Jackson, Creating an anthropomorphic digital MR phantom—an extensible tool for comparing and evaluating quantitative imaging algorithms. Phys. Med. Biol. 2016; 61:974
Figures