2690
BIDScoin: A user-friendly application to convert imaging data to the Brain Imaging Data Structure1Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands, 2Neurobiology Research Unit, Copenhagen University Hospital, Copenhagen, Denmark, 3Center for Multimodal Neuroimaging, National Institute of Mental Health, Bethesda, MD, United States, 4NatMEG, Karolinska Institutet, Stockholm, Sweden
Synopsis
Sharing neuroimaging data is an important development that has the potential to be scaled up with the new Brain Imaging Data Structure (BIDS) standard. Existing tools to converting data to BIDS often require programming skills or are tailored to specific institutes, datasets or data formats. Here we introduce BIDScoin, a cross-platform, flexible, free and open-source converter that provides a graphical user interface to help users finding their way in the BIDS standard, and supports plugins to extend its functionality. We show its design and demonstrate how it can be applied to a downloadable tutorial dataset.
1. Introduction
Pooling shared neuroimaging data is a promising way to study the brain in health and disease. Recently, the Brain Imaging Data Standard (BIDS1) was introduced to facilitate MRI data sharing and has recently been extended to MEG2, EEG3, iEEG4, genetic5, and PET data6. The BIDS standard has moved the burden of homogenizing the heterogeneous data formats and structures in the various sources from the end-user to those that have collected the dataset. A current limitation, however, is that many of those researchers do not possess the programming skills to efficiently reformat their data to BIDS. Various BIDS conversion tools have been made available, but they generally still require programming skills and lack graphical user interfaces (GUI).The user-friendly BIDScoin application presented here can flexibly convert various kinds of source data to BIDS without programming anything. BIDScoin uses an intelligent datatype mapping approach that exploits as much of the digital information about the data as possible, as well as information typically known only by the researcher. These mappings are intuitive for researchers as they (1) resemble the way they often think about their datatypes, (2) are simple and flexible, and (3) come with a GUI to easily edit them to their needs and data knowledge.
2. Method
All BIDScoin Python 3.6 code (https://github.com/Donders-Institute/bidscoin) and documentation (https://bidscoin.readthedocs.io) are freely available.2.1 BIDScoin workflow
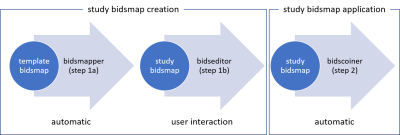
The BIDScoin workflow consists of three different steps (Figure 1):- Data mapping. A “template bidsmap” is used to scan the source dataset and automatically create a “study bidsmap”, containing all the discovered BIDS-mappings from source datatypes to BIDS datatypes (Figure 2). The template bidsmap is generic and can contain so-called BIDS-mappings with regular expressions for intelligent data discovery, the study bidsmap contains BIDS-mappings that are narrowed down to the study data.
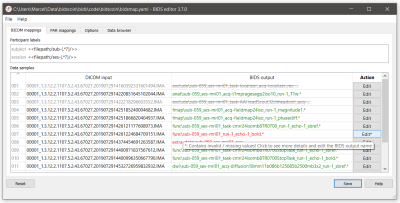
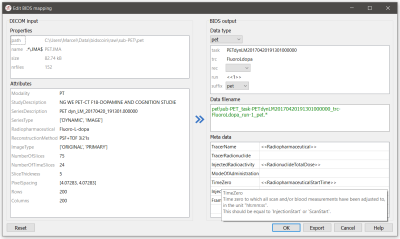
- User interaction. A GUI application can be launched (automatically in step 1a or manually) by the user to edit the suggested BIDS-mappings using their knowledge about the study (Figure 3, 4).
- Data conversion. In this step, the source data is automatically converted (“coined”) to BIDS, as specified in the study bidsmap.
2.2 BIDS-mappings
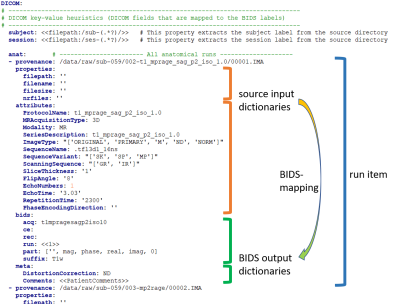
Source data typically comes with two sources of metadata, namely (1) filesystem properties, such as parts of the folder or file name, and (2) header attributes, such as the DICOM “SeriesDescription”. Similarly, metadata is also stored (3) in the BIDS filename and (4) in a sidecar file with metadata. A BIDS-mapping contains dictionaries that map (1) and (2) onto (3) and (4) (see Figure 2). The dictionary keys of input dictionaries (1) and (2) are used to extract metadata from the data that is compared to the associated dictionary values. A BIDS-mapping is established if all dictionary items of (1) and (2) match. Importantly, in the matching procedure, the dictionary values are interpreted as regular expression patterns. In this way, the template bidsmap can contain intelligence / prior knowledge about the data. In the study bidsmap, the regular expression values are replaced (narrowed down) by the extracted metadata values.Users can directly set the BIDS-mapping values of output dictionaries (3) and (4) as they like, but often these values are available as file attributes or properties or they may vary between acquisitions. BIDScoin allows researchers to capture such cases with so-called “dynamic values”, when enclosed by single (<>) or double brackets (<<>>). Then the value is taken as an attribute or property key for which the value is then extracted from the data. Moreover, substrings from dynamic values can be taken by appending a colon-separated regular expression. This allows e.g. researchers to extract multiple values or key-value pairs from a single header attribute or source filename.
2.3 Plugins
Architecturally, to facilitate the implementation of new BIDS developments, all interactions in BIDScoin with source data are done via plugins that have dataformat-independent interfaces. Currently, BIDScoin comes with these pre-installed plugins:- dcm2niix2bids: A plugin that wraps around pydicom7, nibabel8 and dcm2niix9 to handle DICOM and Philips PAR(/REC)/XML source data.
- spec2nii2bids: A plugin that wraps around spec2nii to handle MR spectroscopy data.
- phys2bidscoin: An experimental plugin that wraps around the phys2bids library (https://zenodo.org/record/4983522) to handle physiological labchart (ADInstruments) and AcqKnowledge (BIOPAC) source data.
- pet2bidscoin: An experimental plugin that wraps around pet2bids to handle ECAT and DICOM PET source data.
3 Discussion
We have demonstrated the use-case for and main working of our user-friendly BIDScoin application that can convert a variety of raw neuroimaging data formats to BIDS. Importantly, BIDScoin does not require programming, comes with a GUI, has a flexible plugin architecture, and can be installed on Linux, Windows and macOS. Power users can code their own plugins and use regular expressions to deal with special use cases or extend the functionality of the application.There is still a need to develop more plugins to support more data formats and interface with data management solutions such as DataLad10. Code quality can be improved by increasing the code coverage of the automated tests.
Acknowledgements
We would like to thank Rutger van Deelen for providing the initial (PyQt) setup and implementation of the bidseditor application. We are also grateful for all the feedback, questions and contributions that users have submitted on GitHub.References
1) Gorgolewski, K., Auer, T., Calhoun, V. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data 3, 160044 (2016). https://doi.org/10.1038/sdata.2016.44
2) Niso, G., Gorgolewski, K. J., Bock, E., Brooks, T. L., Flandin, G., Gramfort, A., Henson, R. N., Jas, M., Litvak, V., T Moreau, J., Oostenveld, R., Schoffelen, J. M., Tadel, F., Wexler, J., & Baillet, S. (2018). MEG-BIDS, the brain imaging data structure extended to magnetoencephalography. Scientific data, 5, 180110. https://doi.org/10.1038/sdata.2018.110
3) Pernet, C. R., Appelhoff, S., Gorgolewski, K. J., Flandin, G., Phillips, C., Delorme, A., & Oostenveld, R. (2019). EEG-BIDS, an extension to the brain imaging data structure for electroencephalography. Scientific data, 6(1), 103. https://doi.org/10.1038/s41597-019-0104-8
4) Holdgraf, C., Appelhoff, S., Bickel, S., Bouchard, K., D'Ambrosio, S., David, O., Devinsky, O., Dichter, B., Flinker, A., Foster, B. L., Gorgolewski, K. J., Groen, I., Groppe, D., Gunduz, A., Hamilton, L., Honey, C. J., Jas, M., Knight, R., Lachaux, J. P., Lau, J. C., … Hermes, D. (2019). iEEG-BIDS, extending the Brain Imaging Data Structure specification to human intracranial electrophysiology. Scientific data, 6(1), 102. https://doi.org/10.1038/s41597-019-0105-7
5) Moreau, C. A., Jean-Louis, M., Blair, R., Markiewicz, C. J., Turner, J. A., Calhoun, V. D., Nichols, T. E., & Pernet, C. R. (2020). The genetics-BIDS extension: Easing the search for genetic data associated with human brain imaging. GigaScience, 9(10), giaa104. https://doi.org/10.1093/gigascience/giaa104
6) Knudsen, G.M., Ganz, M., Appelhoff, S., Boellaard, R., Bormans, G., Carson, R.E., Catana, C., Doudet, D., Gee, A.D., Greve, D.N., Gunn, R.N., Halldin, C., Herscovitch, P., Huang, H., Keller, S.H., Lammertsma, A.A., Lanzenberger, R., Liow, J.S., Lohith, T.G., Lubberink, M., Lyoo, C.H., Mann, J.J., Matheson, G.J., Nichols, T.E., Nørgaard, M., Ogden, T., Parsey, R., Pike, V.W., Price, J., Rizzo, G., Rosa-Neto, P., Schain, M., Scott, P.J.H., Searle, G., Slifstein, M., Suhara, T., Talbot, P.S., Thomas, A., Veronese, M., Wong, D.F., Yaqub, M., Zanderigo, F., Zoghbi, S., Innis, R.B. (2020). Guidelines for Content and Format of PET Brain Data in Publi-cations and in Archives: A Consensus Paper. Journal of Cerebral Blood Flow and Metabolism, 2020 Aug; 40(8): 1576-1585. doi:10.1177/0271678X20905433
7) Mason, D., scaramallion, rhaxton, mrbean-bremen, Suever, J., Vanessasaurus, Lemaitre, G., Papadopoulos Orfanos, D., Panchal, A., Rothberg, A., Massich, J., Kerns, J., van Golen, K., Robitaille, T., moloney, Shun-Shin, M., pawelzajdel, Conrad, B., Mattes, M., Biggs, S., Morency, F., Herrmann, M., Meine, H., Wortmann, J., Hahn, K., Wada, M., Rachum, R., colonelfazackerley, ferdymercury, huicpc0207 (2020). Pydicom: An open source DICOM library. doi: 10.5281/zenodo.4313150
8) Brett, M., Markiewicz, C., Hanke, M., Côté, M., Cipollini, B., McCarthy, P., Jarecka, D., Cheng, C., Halchenko, Y., Cottaar, M., Larson, E., Ghosh, S., Wassermann, D., Gerhard, S., Lee, G., Wang, H., Kastman, E., Kaczmarzyk, J., Guidotti, R., Duek, O., Daniel, J., Rokem, A., Madison, C., Moloney, B., Morency, F., Goncalves, M., Markello, R., Riddell, C., Burns, C., Millman, J., Gramfort, A., Leppäkangas, J., Sólon, A., van den Bosch, J., Vincent, R., Braun, H., Subramaniam, K., Gorgolewski, K., Raamana, P., Klug, J., Nichols, B., Baker, E., Hayashi, S., Pinsard, B., Haselgrove, C., Hymers, M., Esteban, O., Koudoro, S., Pérez-García, F., Oosterhof, N., Amirbekian, B., Nimmo-Smith, I., Nguyen, L., Reddigari, S., St-Jean, S., Panfilov, E., Garyfallidis, E., Varoquaux, G., Legarreta, J., Hahn, K., Hinds, O., Fauber, B., Poline, J., Stutters, J., Jordan, K., Cieslak, M., Moreno, M., Haenel, V., Schwartz, Y., Baratz, Z., Darwin, B., Thirion, B., Gauthier, C., Papadopoulos Orfanos, D., Solovey, I., Gonzalez, I., Palasubramaniam, J., Lecher, J., Leinweber, K., Raktivan, K., Calábková, M., Fischer, P., Gervais, P., Gadde, S., Ballinger, T., Roos, T., Reddam, V., freec84 (2020). Python package to access a cacophony of neuro-imaging file formats. doi: 10.5281/zenodo.4295521
9) Li, X., Morgan, P.S., Ashburner, J., Smith, J., Rorden, C. (2016) The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J Neurosci Methods. 264:47-56.
10) Halchenko, Y., Meyer, K., Poldrack, B., Solanky, D., Wagner, A., Gors, J., Macfarlane, D., Pustina, D., Sochat, V., Ghosh, S., Mönch, C., Markiewicz, C., Waite, L., Shlyakhter, I., Vega, A., Hayashi, S., Häusler, C., Poline, J., Kadelka, T., Hanke, M. (2021). DataLad: distributed system for joint management of code, data, and their relationship. Journal of Open Source Software 6: 3262. doi: 10.21105/joss.03262
Figures