2678
MR imaging of Zebrafish with mitochondria diseases
Sergey Magnitsky1, Sonal Sharma1, and Marni Falk1
1CHOP, Philadelphia, PA, United States
1CHOP, Philadelphia, PA, United States
Synopsis
We created a high-resolution MR imaging atlas of wt. and three mutants (OPA1, FBXL4
Introduction
Zebrafish are vertebrates with many advantages over traditional mammalian models, including rapid development, large brood size, and ease of genetic manipulation. Zebrafish models are used extensively for the development of new therapeutic approaches to mitochondria disorders. This animal model provides an excellent representation of abnormal development, demyelination, cardiomyopathy, hepatomegaly and tissue atrophy. This study aimed to create an anatomical atlas of normal and clinically relevant mutants of Zebrafish based on high-resolution MR imaging. We analyzed wt. and OPA1, FBXL4 and SURF1 Zebrafish mutants and compared organs' size of these animals.Methods
Animal model: Three (OPA1 (n=3), FBXL4(n=3) and SURF1 (n=9)) Zebrafish mutant and wt.(n=16) animals were reviewed in our experiment. Animals were divided into three age groups: 5, 12-14, 16-18, 31 months old, and organs volume was measured based on MR images.Imaging: MR imaging was performed on a 9.4 T vertical bore Bruker NMR magnet equipped with a 100 Gauss/cm gradient insert for imaging. The experimental animals were sacrificed and incubated in PFA and 2 mM Gd-DTPA for 5 days. Specimens were placed in a 10 mm NMR tube with 4 ml of Fomblin. 3D gradient-echo images of the fish specimens were acquired with the following parameters TR=100 ms, TE = 6 ms, flip angle 800 with final isotropic 30, 50, 75 mm3 resolution.
Image analysis: Volume of liver, heart, eyes, optic nerve, kidneys, swim bladder, brain, ears, spinal cord were calculated after delineation of these organs using ITK-SNAP software. A linear regression method was implemented to examine the dependence of the organs volume and age of the animals. T-test was implemented to delineate a difference between organ size.
Results
Figure 1 shows MR images of wt. Zebrafish acquired at a different resolution. High-resolution 30 mm3images were selected for characterization of mutant specimens.We investigated organ size as a function of the age of the experimental animals. Liner increase of organs volume with aging was detected for all tested animals. Representative 3D rendering of different organs is shown in Figure 2.
Figure 3 depicts organ size in wt. and mutants.
Table 1 presents the P-values of wt. and different
Discussion and conclusions
Our long-term goal is to develop an MRI platform for in vivo investigation of Zebrafish models of human diseases. In this study, we were interested in the anatomical characterization of wt. Zebrafish with OPA1, FBXL4 and SURF1 mutations. These mutations were found in patients with mitochondrial disorders and Zebrafish models were created for preclinical experiments. Anatomical and functional characterization of these models is critically important for the justification of Zebrafish models and a development of noninvasive imaging markers for testing new therapies. Histological staining is a standard procedure for a description of animal models. However, this technique becomes not practical for evaluating a large amount of animal models with a single gene nutation due to excesses slicing and staining work. In our experiments, we demonstrated that high-resolution 3D MRI provides comparable with histology resolution, allows to observe and evaluate the entire animal and does not require an enormous amount of work as histological staining. In this study we demonstrated that OPA1 mutant exhibits smaller brain and spinal cord among three different mutants compared to wt. animals.Acknowledgements
We would like to thank Dr. C. Seiler from the CHOP Zebrafish core facility for support with animal modes.References
No reference found.Figures
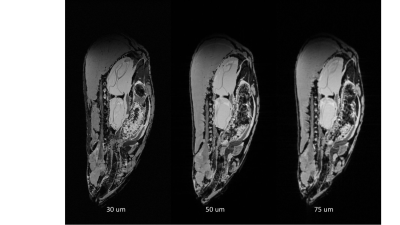
MR image of wt. Zebrafish acquired with 30, 50 and 75 mm3 resolution
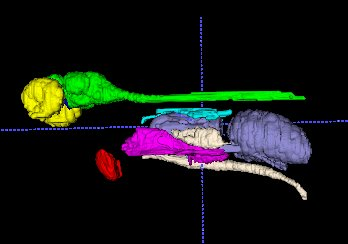
3D model of eyes (yellow), optic nerve (blue), Brain and spinal cord (green), heart (red), liver (purple) kidney (light blue), swim bladder (dark blue) and testis (white) were created based on the MR image of wt. Zebrafish.
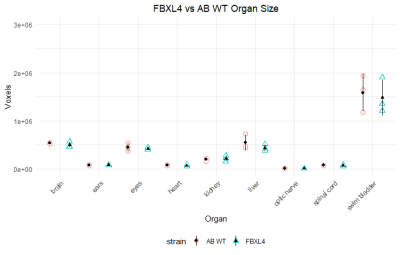
FBXL4 mutant
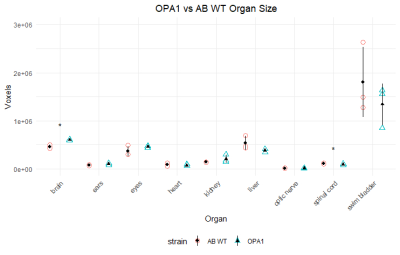
OPA1 mutant
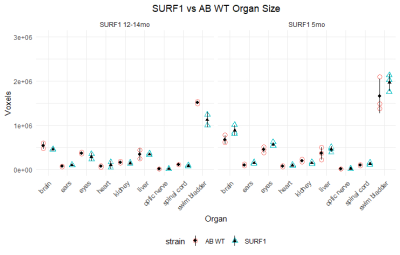
SURF1 mutant
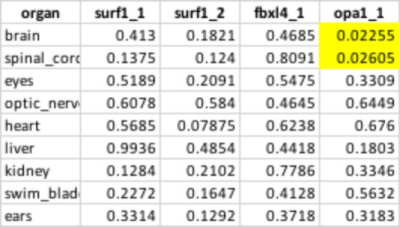
Table 1 P-values of wt. and different mutants.
DOI: https://doi.org/10.58530/2022/2678