2654
Automated machine learning-based brain lesion segmentation on structural MRI acquired from traumatic brain injury patients1Keck School of Medicine, University of Southern California, Los Angeles, CA, United States, 2David Geffen School of Medicine, University of California, Los Angeles, LOS ANGELES, CA, United States
Synopsis
Traumatic brain injury (TBI) can cause severe disorders, including post-traumatic epilepsy (PTE). Lesion segmentation is an MRI-based analysis to identify brain structures that correlate with PTE development post-TBI. Unfortunately, manual segmentation, considered the gold standard, is highly tedious and noisy. Thus, we propose the first automated machine-learning based lesion segmentation method for MRI of TBI patients enrolled in the Epilepsy Bioinformatics Study for Antiepileptogenic Therapy (EpiBioS4Rx). Experimental validation demonstrates considerable visual overlap of lesion predictions and ground-truths with 61% precision. Early and automated lesion segmentation via our approach can aid experts in MRI analysis and successful PTE identification following TBI.
Abstract Body
Traumatic brain injury (TBI) is a prevalent cause of severe disorders, such as post-traumatic epilepsy (PTE). PTE is diagnosed if unprovoked recurrent seizures occur at least one week post-TBI, though they can emerge several years after a TBI[1]. The unpredictable onset of PTE motivates early identification of epileptic biomarkers. The Epilepsy Bioinformatics Study for Antiepileptogenic Therapy (EpiBioS4Rx) is a multi-site project with the goal of identifying biomarkers of epileptogenesis after a TBI via the collection and analysis of large-scale multimodal data[2]. Magnetic resonance imaging (MRI), with particular focus on structural MRI (sMRI), has been commonly employed due to its non-invasive nature and high visual resolution[3,4].TBI characteristics such as lesion type, location, and extent have been shown to be correlated with increased risk of PTE[5], motivating the use of lesion phenotype analysis as a means for early PTE identification. An important MRI-based analysis in this regard is parenchymal lesion segmentation, which is a process to identify relevant lesion regions or structures affected by hemorrhagic contusion or edema within MRI. Unfortunately, manual execution of segmentation on medical images such as MRI not only is a highly tedious and time-consuming task, but also exhibit inter-rater variability for clinical experts[6,7,8]; this motivates the need for automated segmentation tools to support the long-lasting efforts of clinicians and improve visual explainability of diagnostic decisions. In the last decade, the rapid development of automated segmentation algorithms is strongly connected with the rising success and popularity of machine learning. Particularly, a specific field of machine learning, termed as a deep neural network (DNN)[9], has outperformed traditional segmentation methods.
Motivated by these observations, we propose the first DNN architecture that automatically segments lesions on sMRI acquired from TBI patients. Our approach is inspired by a state-of-the-art DNN architecture which automatically segments brain tumour regions on MRI[10]. The architecture receives an MRI scan and predicts each voxel as belonging to a lesion or background, as depicted in Figure 1. We validate the automated segmentation performance of the DNN architecture on a dataset of T2-weighted Fluid-Attenuated Inversion Recovery (FLAIR) sMRI scans acquired from 67 patients enrolled in EpiBioS4Rx. Each sMRI scan is annotated with a ground-truth lesion region using the ITK-SNAP tool[11], validated by a physician with neuroradiological expertise, and prepared by registration with respect to a Montreal Neurological Institute (MNI) template. The annotated sMRI dataset is partitioned into training and validation sets, allocating 53 scans for training and 14 scans for validation. We train the DNN architecture using the training scans, with the objective of maximizing the overlap of predicted and ground-truth lesion regions.
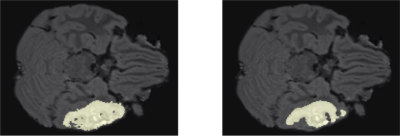
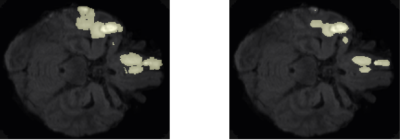
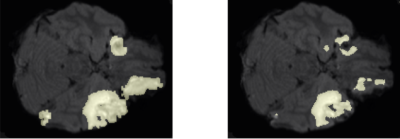
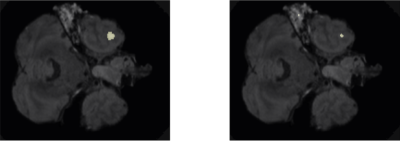
Following training, we qualitatively evaluate the automated lesion segmentation predictions by visualizing ground-truth vs. predicted lesion regions in Figures 2-5 via example slices extracted from MRI scans of several patients. Our method makes considerably similar lesion segment predictions compared to the ground-truth lesions visually, even for the cases of multiple lesion regions (c.f. Figures 3-4) or very small lesions (c.f. Figure 5). Moreover, we quantitatively evaluate the automated lesion segmentation predictions on the validation set scans using Dice similarity, precision, and recall metrics. Dice similarity is a benchmark metric that evaluates the overlap of predicted and ground-truth lesion regions[12], while precision and recall are computed for correctly identifying each voxel as lesion vs. background and are averaged over all voxels. Our DNN architecture attains 49% Dice similarity score, 61% precision, and 49% recall over the validation set. As shown in Figures 2-5, our architecture tends to underestimate lesion volumes, which is reflected in the quantitative performance averaged over the validation set. We also observe that segmentation predictions are affected by the quality of registration prior to automated segmentation, as traditional software tools for registration are not optimized for traumatic brains[4]. Overall, the considerable visual similarities of predicted and ground-truth lesion regions, along with the quantitative evaluations, illustrate the potential success of our DNN approach in automated lesion segmentation.
In this work, we take the first successful steps of automated machine learning-based brain lesion segmentation on sMRI acquired from TBI patients. Future work is planned to improve automated segmentation performance via better image registration and tailoring the training objective for underestimated volumes. Early and automated segmentation of brain lesions observed on TBI patients can aid successful PTE and epileptogenesis identification by indicating clinically relevant brain regions and structures. This type of visual aid in MRI analysis can in turn significantly help clinicians save time and effort and facilitate successful clinical trials of antiepileptogenic therapy.
Acknowledgements
Our work is supported by NIH Grants NINDS U54 NS100064, and is disseminated on behalf of The Epilepsy Bioinformatics Study for Antiepileptogenic Therapy (EpiBioS4Rx) investigators.References
- Diaz-Arrastia, R., Agostini, M. A., Madden, C. J., and Van Ness, P. C. (2009). Posttraumatic epilepsy: the endophenotypes of a human model of epileptogenesis. Epilepsia 50, 14–20. doi: 10.1111/j.1528-1167.2008.02006.x
- Vespa, P.M., Shrestha, V., Abend, N., Agoston, D., Au, A., Bell, M.J., Bleck, T.P., Blanco, M.B., Claassen, J., Diaz-Arrastia, R. and Duncan, D., 2019. The epilepsy bioinformatics study for anti-epileptogenic therapy (EpiBioS4Rx) clinical biomarker: study design and protocol. Neurobiology of disease, 123, pp.110-114.
- Lutkenhoff ES, Shrestha V, Tejeda JR, Real C, McArthur DL, Duncan D, La Rocca M, Garner R, Toga AW, Vespa PM, Monti MM, Early brain biomarkers of post-traumatic epilepsy: initial report of the multicenter Epilepsy Bioinformatics Study for Anti-epileptogenic Therapy (EpiBioS4Rx) prospective study, Journal of Neurology, Neurosurgery, and Psychiatry. Aug 26 2020.
- La Rocca M, Garner R, Lutkenhoff ES, Monti MM, Amoroso N, Vespa P, Toga AW, Duncan D, Multiplex networks to characterize seizure development in traumatic brain injury patients, Frontiers in Neuroscience. November 2020
- Garner R, La Rocca M, Vespa P, Jones N, Monti MM, Toga AW, Duncan D, Imaging biomarkers of posttraumatic epileptogenesis, Epilepsia. October 2019.
- Norouzi, A., Rahim, M.S.M., Altameem, A., Saba, T., Rad, A.E., Rehman, A. and Uddin, M., 2014. Medical image segmentation methods, algorithms, and applications. IETE Technical Review, 31(3), pp.199-213.
- Renard, F., Guedria, S., De Palma, N. and Vuillerme, N., 2020. Variability and reproducibility in deep learning for medical image segmentation. Scientific Reports, 10(1), pp.1-16.
- Sharma, N. and Aggarwal, L.M., 2010. Automated medical image segmentation techniques. Journal of medical physics/Association of Medical Physicists of India, 35(1), p.3.
- Goodfellow, I., Bengio, Y. and Courville, A., 2016. Deep learning. MIT press.
- Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T. and Ronneberger, O., 2016, October. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In International conference on medical image computing and computer-assisted intervention (pp. 424-432). Springer, Cham.
- Paul A. Yushkevich, Joseph Piven, Heather Cody Hazlett, Rachel Gimpel Smith, Sean Ho, James C. Gee, and Guido Gerig. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006 Jul 1;31(3):1116-28.
- Milletari, F., Navab, N. and Ahmadi, S.A., 2016, October. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In 2016 fourth international conference on 3D vision (3DV) (pp. 565-571). IEEE.
Figures