2651
Machine Learning-Based Radiomics Predicting the IDH1 Genotype of Diffuse Gliomas1Department of Radiology, The First Affiliated Hospital, Kunming Medical University, Kunming, China, 2Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou, China, 3Fisca Healthcare Ltd, Kunming, China, 4Department of Oncologic Imaging, National Cancer Center, Singapore, Singapore, 5GE Healthcare, MR Research, Beijing, China
Synopsis
The current study aims to evaluate the value of susceptibility weighted imaging (SWI) and contrast-enhanced T1-weighted imaging (CE-T1WI) radiomics features in predicting isocitrate dehydrogenase1 (IDH1) genotype of diffuse gliomas and build prediction models. It was concluded that SWI and CE-T1WI radiomics features can effectively predict the IDH1 genotype of diffuse gliomas, and CE-T1WI performed better. By combining SWI with CE-T1WI radiomics features, the prediction performance can be improved.
Introduction
Studies have shown that IDH1 genotype is a critical biomarker for precision diagnosis, prognosis prediction and a potential treatment target [1, 2]. At present, IDH1 genotype is mainly identified by immunohistochemistry or gene sequencing after biopsy or surgical resection, which are invasive procedures and prone to inaccurate pathological classification, because of the heterogeneity of diffuse glioma. Fortunately, studies found that radiomics can quantitatively depict the tumor characteristics with high throughput mathematical features extracted from medical imaging data, and establish a predictive model to evaluate the tumor genotype through relevant features noninvasively [3]. Images of CE-T1WI contain necrosis and the integrity of the blood-brain barrier information in the tumor, and SWI can reflect the intratumoral microhemorrhage and neoangiogenesis [4], providing complement information from CE-T1WI regarding intratumoral heterogeneous components. This study is to build IDH1 genotype prediction models with selected radiomics features from images of SWI and CE-T1WI and evaluate the performance of each model.Material and Methods
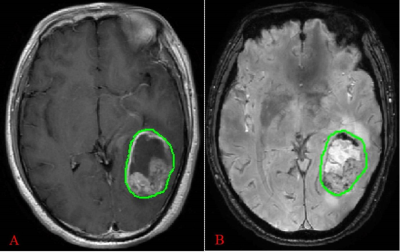
An analysis was performed on 50 patients with pathology confirmed diffuse glioma, including 16 cases of IDH1-mt (7 males and 9 females,42.7 ± 10.4 years old ) and 34 cases of IDH1-wt (16 males and 18 females,52.0 ± 10.9 years old ). All patients underwent examination on GE 3.0 T MRI scanner (Discovery MR750, GE, US) preoperatively. Standard MR imaging protocol includes T2WI, T1WI, T2-FLAIR, SWI and CE-T1WI. Using IBEX software to delineate region of interest (ROI) on the SWI magnitude map and axial CE-T1WI (Figure 1), then automatically analyze and extract radiomics features. Feature selection was performed by F-test, mutual information (MI), minimum redundancy maximum relevance (MRMR) and least absolute shrinkage and selection operator (LASSO) Cox regression model. Prediction models were built with selected features and were trained and validated using leave-one-out cross validation (LOOCV). Receiver operating characteristic (ROC) curve and area under the curve (AUC) were used to evaluate the prediction efficiency of each model.Results
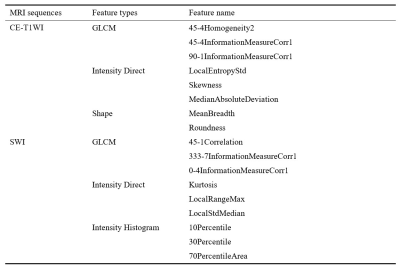
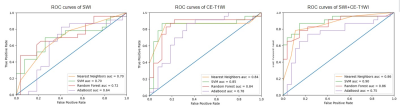
Nine SWI radiomics features and eight CE-T1WI radiomics features were selected. The selected radiomics features are summarized in Table 1. The overall models built with selected radiomics features achieved a predictive performance. Models based on the CE-T1WI performed better than that of SWI. By combining SWI with CE-T1WI radiomics features, the prediction performance can be improved. Additionally, Models with support vector machines (SVM) were found to be consistently performed best in all models, and achieved the best predictive performance (SWI, AUC = 0.70; CE-T1WI, AUC = 0.85, SWI+CE-T1WI, AUC = 0.90). The ROC curves of the radiomics models are shown in Figure 2.Discussion and Conclusion
Studies have shown that some radiomics features and IDH1 genotype are closely related [5, 6]. In this study, the radiomics features based on SWI and CE-T1WI images could predict IDH genotype in diffuse gliomas, mainly including GLCM, intensity direct, intensity histogram and shape. The intensity direct and GLCM features account for a large proportion, this is consistent with previous studies. We evaluated the ability of radiomics features from individual MRI sequence to predict IDH1 genotype, all models can effectively predict the IDH1 genotype of diffuse gliomas, and CE-T1WI performed better than SWI. The one reason might be that the image resolution of the SWI sequence was lower than the CE-T1WI images, which influenced the stability and robustness of the radiomics features. The other might be that the SWI sequence was affected by susceptibility artifacts in the air or bone tissue interface, thereby influencing tumors located in these regions. However, SWI can reflect intratumoral microhemorrhage and neoangiogenesis without administration of an exogenous contrast agent, so SWI sequence is promising in predicting IDH1 genotype.It has been shown that prediction performance can be improved by combining different sequences [7, 8]. This study showed that the prediction performance of two sequences combined was higher than that of an individual sequence over all models, which was concordant with previous findings. This suggests that the radiomics features of different sequences reflect a part of the imaging features expressed by IDH1 and can complement each other.Acknowledgements
This work was supported by Yunnan Provincial Science and Technology Department, Kunming Medical University applied basic research (2019FE001(-052)), and Yunnan Provincial Health Science and Technology Program (2018NS0120).References
1. Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743-50.
2. Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120(6):707-18.
3. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441-6.
4. Thompson EM, Keir ST, Venkatraman T, Lascola C, Yeom KW, Nixon AB, et al. The role of angiogenesis in Group 3 medulloblastoma pathogenesis and survival. Neuro Oncol. 2017;19(9):1217-27.
5. Yu J, Shi Z, Lian Y, Li Z, Liu T, Gao Y, et al. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol. 2017;27(8):3509-22.
6. Ren Y, Zhang X, Rui W, Pang H, Qiu T, Wang J, et al. Noninvasive Prediction of IDH1 Mutation and ATRX Expression Loss in Low-Grade Gliomas Using Multiparametric MR Radiomic Features. J Magn Reson Imaging. 2019;49(3):808-17.
7. Tan Y, Zhang S-t, Wei J-w, Dong D, Wang X-c, Yang G-q, et al. A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. European Radiology. 2019;29(7):3325-37.
8. Peng H, Huo J, Li B, Cui Y, Zhang H, Zhang L, et al. Predicting Isocitrate Dehydrogenase (IDH) Mutation Status in Gliomas Using Multiparameter MRI Radiomics Features. J Magn Reson Imaging. 2021;53(5):1399-407.