2614
Harmonisation of multi-site MRS data with ComBat
Tiffany Bell1,2,3, Kate J Godfrey1,2,3, Ashley L Ware2,3,4,5, Keith Owen Yeates2,3,4, and Ashley D Harris1,2,3
1Department of Radiology, University of Calgary, Calgary, AB, Canada, 2Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 3Alberta Children's Hospital Research Institute, University of Calgary, Calgary, AB, Canada, 4Department of Psychology, University of Calgary, Calgary, AB, Canada, 5Department of Neurology, University of Utah, Salt Lake City, UT, United States
1Department of Radiology, University of Calgary, Calgary, AB, Canada, 2Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 3Alberta Children's Hospital Research Institute, University of Calgary, Calgary, AB, Canada, 4Department of Psychology, University of Calgary, Calgary, AB, Canada, 5Department of Neurology, University of Utah, Salt Lake City, UT, United States
Synopsis
Multisite magnetic resonance spectroscopy (MRS) studies are becoming increasingly more common, however data collection across multiple sites introduces non-biological variability that can mask true biological effects. Using PRESS and MEGA-PRESS data obtained from the BIG GABA repository and linear modelling, we show that (1) scanner vendor and data collection site is significantly associated with metabolite levels and (2) data harmonisation using ComBat successfully removes non-biological variance to reveal biological effects of interest. We therefore recommend ComBat as an approach to harmonise multi-site MRS data.
Introduction
Magnetic resonance spectroscopy (MRS) is a non-invasive neuroimaging technique that measures brain chemistry in vivo and has been used to study the healthy brain as well as numerous neurological disorders. Multi-site MRS studies are increasing, however, non-biological variability introduced during data collection across multiple locations (e.g., due to differences across scanner vendors and site-specific acquisition implementations) can hinder detection of biological effects of interest.Indeed, Mikkelsen et al. showed significant effects of both vendor and site on GABA+ and a significant effect of site on macromolecule-suppressed GABA levels both when referencing to creatine1 and water.2 Považan et al.3 found similar effects of vendor and site on PRESS data.
ComBat is a data harmonisation technique that can remove unwanted variability caused by differences in data collection location while preserving true biological variability. ComBat has been validated for harmonizing structural and functional MRI metrics, but not yet for MRS metabolite measures. This study aimed to validate ComBat harmonisation for single voxel MRS data in healthy adults.
Methods
Data were obtained from the BIG GABA repository (www.nitrc.org/projects/biggaba/), which consists of single voxel MRS data acquired from a 3x3x3 cm3 voxel in the parietal lobe in adults aged 18-35. Data were acquired on 3T MRI scanners from three major vendors: General Electric (GE), Philips, and Siemens.1 PRESS data (N=195; TR/TE=2000/35 ms) were included from 19 sites. GABA+ (N=218, TR/TE=2000/68 ms, 15 ms editing pulses at 1.9/7.5 ppm) and macromolecule-suppressed GABA (N=209; GE and Phillips: TR/TE=2000/80 ms, Siemens: TR/TE=2000/68 ms, 20 ms editing pulses at 1.9/1.5 ppm) data were included from 20 sites.PRESS data were quantified using LCModel4 with basis sets simulated using FID-A.5 PRESS metabolites included in the harmonisation analysis were tNAA (total NAA, NAA+NAAG), tCr (total creatine, creatine+phosphocreatine), tCho (total choline, glycerophosphocholine+phosphocholine), Glx (glutamate+glutamine) and Myo-inositol (Myo-Ins). MEGA-PRESS data were preprocessed and quantified using Gannet 3.2.6 All data were quantified in molal units and corrected for partial volume effects.7 MEGA-PRESS data quantification included the α-correction, which assumes twice as much GABA in grey vs white matter.8 A secondary, confirmatory analysis for all metabolites (except tCr) was performed using tCr-referenced data.
Data were harmonised using neuroComBat version 1.0.5 (https://github.com/Jfortin1/ComBatHarmonization/tree/master/R), implemented in R version 4.0.3 (R Core Team (2020) https://www.R-project.org/). A matrix including the biological covariates of interest (grey matter fraction [fGM/(fGM+fWM)], age and sex) was included to preserve biological variability.
For each metabolite, vendor and site level effects were assessed in three-level unconditional linear mixed-effects models, with vendor and site included as random effects, using the R lme4 package (version 1.1-26 ).10 To validate that ComBat harmonisation maintains biological effects, secondary multilevel analyses were performed to test for effects of grey-matter fraction, age, and sex on each metabolite level.
Results
Evaluation of ComBat HarmonisationWater-referenced data: Prior to harmonization, vendor and site were significantly associated with all metabolites, except macromolecule-suppressed GABA, which showed significant effects of vendor but not site. After harmonisation by site, neither vendor nor site effects were significant (Figure 1; Figure 2).
tCr-referenced data: Prior to harmonisation, there was a significant effect of vendor, but not site. Thus, data were initially harmonised by vendor. However, the effect of vendor remained significant in the model, so data was therefore harmonised by site. After harmonisation by site, no vendor or site effects were significant.
Biological Variability
Water-referenced data: Prior to harmonisation, grey matter fraction, age, and sex were not significantly associated with metabolite levels. Following harmonisation, grey matter fraction was significantly associated with GABA+ levels and sex was significantly associated with tCho levels (males>females).
tCr-referenced data: Sex was significantly associated with tCho/tCr levels both prior to and following harmonisation (males>females). No other biological variables were associated with metabolite levels prior to harmonisation. Following harmonisation, age was significantly associated with GABA+/tCr and Glx/tCr levels, with both metabolite levels decreasing with age.
Discussion
Linear modelling showed significant effects of vendor and site on metabolite levels. Following ComBat harmonisation by site, neither site nor vendor effects were significant (whether referencing to water or tCr), indicating ComBat harmonisation by site successfully removed this non-biological variance. Interestingly, harmonisation by vendor did not completely remove variability across vendors for all metabolites. This could result from the nested nature of vendor within site, such that variance from each variable will be highly overlapped. We therefore recommend harmonisation by the lowest source of variance, in this case site.Harmonisation of MRS data revealed biological effects that were previously not detected, even when controlling for vendor and site in the model, indicating that (1) ComBat harmonisation does remove non-biological variance to allow detection of biological effects and (2) controlling for these factors in statistical models is not enough to remove unwanted variance. These effects are consistent with the literature and confirm the utility of the harmonisation procedures.
Conclusion
We show that ComBat is an effective approach to harmonise MRS data (both PRESS and MEGA-PRESS), and successfully removes non-biological variance due to site and vendor differences, allowing the detection of expected biological effects of interest. We therefore recommend ComBat for harmonisation of MRS data in multi-site studies.Acknowledgements
This work was undertaken thanks in part to funding from: The Natural Sciences and Engineering Research Council of Canada, T. Chen Fong Postdoctoral Fellowship in Medical Imaging (TB), Harley N. Hotchkiss- Samuel Weiss Postdoctoral Fellowship (AW), Killam Postdoctoral Fellowship (AW), Harley N. Hotchkiss doctoral Fellowship (KG) and supported by the Hotchkiss Brain Institute and the Alberta Children’s Hospital Research Institute, University of Calgary. Data collection was undertaken thanks to NIH grant EB016089. ADH holds a Canada Research Chair in Magnetic Resonance Spectroscopy in Brain Injury and KOY holds a Ronald and Irene Ward Chair in Pediatric Brain Injury.References
- Mikkelsen, M. et al. Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage 159, 32–45 (2017).
- Mikkelsen, M. et al. Big GABA II: Water-Referenced Edited MR Spectroscopy at 25 Research Sites. Physiol. Behav. 176, 139–148 (2019).
- Považan, M. et al. Comparison of multivendor single-voxel MR spectroscopy data acquired in healthy brain at 26 sites. Radiology 295, 171–180 (2020).
- Provencher, S. W. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed 14, 260–264 (2001).
- Simpson, R., Devenyi, G. A., Jezzard, P., Hennessy, T. J. & Near, J. Advanced processing and simulation of MRS data using the FID appliance (FID-A)—An open source, MATLAB-based toolkit. Magn. Reson. Med. 77, 23–33 (2017).
- Edden, R. A. E., Puts, N. A. J., Harris, A. D., Barker, P. B. & Evans, C. J. Gannet: A Batch-Processing Tool for the Quantitative Analysis of Gamma-Aminobutyric Acid–Edited MR Spectroscopy Spectra. J Magn Reson Imaging 40, 1445–1452 (2014).
- Near, J. et al. Preprocessing, analysis and quantification in single‐voxel magnetic resonance spectroscopy: experts’ consensus recommendations. NMR Biomed. e4257 (2020) doi:10.1002/nbm.4257.
- Harris, A. D., Puts, N. A. J. & Edden, R. A. E. Tissue correction for GABA-edited MRS: Considerations of voxel composition, tissue segmentation, and tissue relaxations. J. Magn. Reson. Imaging 42, 1431–1440 (2015).
- R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
- Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 1, 1–48 (2015).
Figures
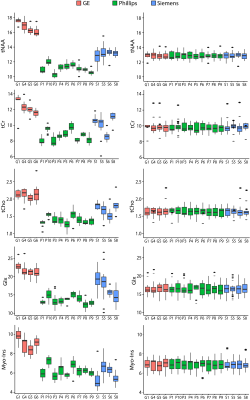
Figure 1: PRESS metabolite levels (referenced to water, units: mol/kg) pre and post-harmonisation. Box plots show median and interquartile range (IQR), dots represent outliers (more that 1.5xIQR).
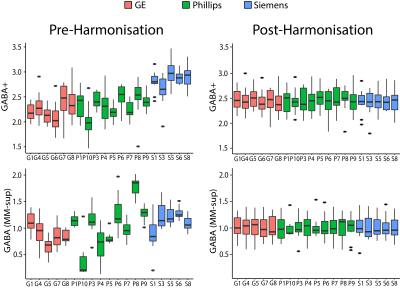
Figure 2: MEGA-PRESS metabolite levels (referenced to water, units: mol/kg) pre and post-harmonisation. Box plots show median and interquartile range (IQR), dots represent outliers (more that 1.5xIQR).
DOI: https://doi.org/10.58530/2022/2614