2579
Automatic 3D bladder segmentation from low-field MR images using 3D U-Net1Magnetic Detection & Imaging, TechMed Centre, University of Twente, Enschede, Netherlands, 2Applied Mathematics, TechMed Centre, University of Twente, Enschede, Netherlands
Synopsis
Pelvic organ prolapse (POP) is a common problem in women, but little is known about treatments. Automatic 3D segmentation of pelvic organs would be useful for improving research in this area. This study successfully applies a 3D U-Net for automatic bladder segmentation of upright and supine low-field MRI scans from asymptomatic women. The resulting network will probably also perform well on data from POP patients. Further improvements are expected when the training data is completed. Future work will focus on segmentation of additional pelvic organs.
Introduction
Pelvic organ prolapse (POP) is a common problem in women1. Several treatments exist, but these often result in recurrences.2,3 Therefore, we perform research to gain more insight in POP, using low-field MRI4. Currently, POP is quantified in 2D MR images, but this is not sufficient to analyze the complexity of this pelvic pathology.Additional information on organ position and orientation can be obtained from 3D segmentation, as shown in Figure 1. Furthermore, 3D organ segmentation is useful for surgical planning and finite element simulation for biomechanical analysis of POP5.
Since manual segmentation is time-consuming (± 6 hours for Figure 1), an automatic method is desired. Research by Feng et al.6 has shown promising results for automatic segmentation of pelvic organs with blurry boundaries from 1.5 tesla (T) images, scanned in supine position. Upright MRI scanning provides better insight into the true degree of prolapse7, but this is only possible with low-field MRI, resulting in lower image quality. Moreover, the variety in bladder shapes makes automatic segmentation challenging.
This study investigates the application of a 3D U-Net for automatic segmentation of pelvic organs from 0.25T supine and upright MRI. This project focuses on the segmentation of the bladder, which is a relevant marker for POP as it is prolapsed in most POP patients.
Methods
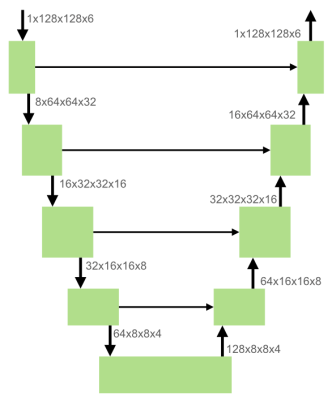
In order to assess the healthy anatomic situation, the pelvic organ position was first analyzed in asymptomatic subjects. The study included 44 women without POP symptoms, between 19 and 65 years of age. Subjects were scanned in upright and supine position using a 0.25T MRI scanner (G-scan Brio, Esaote SpA, Italy) with a 3D bSSFP sequence (TE/TR: 4/8 ms, flip angle: 60°, acquired resolution: 2.02x2.02x2.5 mm³, FOV: 250x250x122 mm³, total scan time: 5:02 min). Images in both body positions are acquired in the same image orientation.In total, 88 scans were made (one supine and one upright scan per participant). Manual annotations of the bladder were obtained in 58 scans by a single expert using 3D Slicer (v.4.11.2021-02-26). These data were randomly divided into a training (24 scans), validation (18 scans), and test set (16 scans), ensuring that scans from a single subject were in the same set. The proposed architecture is a modified 3D U-Net8 (Figure 2), implemented with MONAI9. The network was optimized using an Adam optimizer with Dice loss, a learning rate of 5e-4 and a weight decay of 1e-6. We used early stopping during training, which resulted in 658 epochs.
For evaluation of our network, we used the Dice Similarity Coefficient (DSC) and Hausdorff Distance (HD) in mm. The resulting network was also applied to a scan from an asymptomatic subject whose bladder had prolapsed to emulate the future performance of the network on scans of POP patients.
Results & Discussion
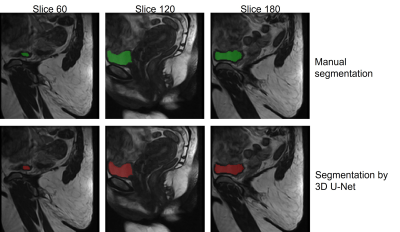
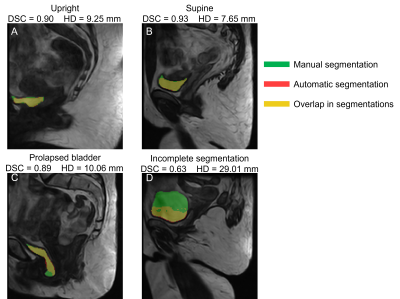
Table 1 shows the performance metrics of the 3D U-Net on the validation and test datasets. Figure 3 shows that the manual segmentation and the automatic segmentation by U-Net in three slices of a 3D upright scan from a single woman correctly align (DSC=0.93, HD=12.09 mm). These results show that bladder segmentation from 3D low-field MRI is promising.Figure 4 shows the scans of four different situations (upright, supine, prolapsed, and unsuccessful) overlayed with the manual and automatic segmentation. Figure 4a and 4b show that segmentation is successful in both supine and upright positions. Furthermore, Figure 4c indicates successful automatic segmentation of a prolapsed bladder in an asymptomatic woman. This suggests that the resulting network will also be applicable to scans from POP patients, with little or even without additional training on POP scans. Moreover, segmentation time was greatly reduced, from 10-60 minutes for a manual bladder segmentation, to only a few seconds by U-Net.
Figure 4d shows an example of incomplete segmentation. In the near future, we will extend our training set with additional annotated images, what will probably improve the robustness of the model.
Future research will focus on the automatic segmentation of additional pelvic organs (uterus, rectum, os pubis, spine) with the same model to further facilitate pelvic research on, e.g., prolapse, pessary positioning and before and after POP treatment comparisons.
Conclusion
This research shows promising results for automatic segmentation of the bladder from low-field upright and supine 3D MRI in asymptomatic women, leading to an enormous reduction in segmentation time. This allows comparing bladder position before and after treatment in 3D, which is useful for research on improving therapy for POP.Acknowledgements
No acknowledgement found.References
1. Slieker-ten Hove, M. C. P. et al. Symptomatic pelvic organ prolapse and possible risk factors in a general population. Am. J. Obstet. Gynecol. 200, 184.e1-184.e7 (2009).
2. Sarma, S., Ying, T. & Moore, K. H. Long-term vaginal ring pessary use: discontinuation rates and adverse events. BJOG An Int. J. Obstet. Gynaecol. 116, 1715–1721 (2009).
3. Fialkow, M. F., Newton, K. M. & Weiss, N. S. Incidence of recurrent pelvic organ prolapse 10 years following primary surgical management: a retrospective cohort study. Int. Urogynecol. J. 19, 1483–1487 (2008).
4. Morsinkhof, L. M., Grob, A. T. M., Veenstra van Nieuwenhoven, A. L. & Simonis, F. F. J. Upright MR imaging of daily variations in pelvic organ position for assessment of pelvic organ prolapse. Proc. Intl. Soc. Mag. Reson. Med. 29 3691, (2021).
5. Chen, L., Ashton-Miller, J. A. & DeLancey, J. O. L. A 3D finite element model of anterior vaginal wall support to evaluate mechanisms underlying cystocele formation. J. Biomech. 42, 1371–1377 (2009).
6. Feng, F., Ashton-Miller, J. A., DeLancey, J. O. L. & Luo, J. Convolutional neural network-based pelvic floor structure segmentation using magnetic resonance imaging in pelvic organ prolapse. Med. Phys. 47, 4281–4293 (2020).
7. Grob, A. T. M. et al. Underestimation of pelvic organ prolapse in the supine straining position, based on magnetic resonance imaging findings. Int. Urogynecol. J. 30, 1939–1944 (2019).
8. Kerfoot, E. et al. Left-Ventricle Quantification Using Residual U-Net BT - Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges. in (eds. Pop, M. et al.) 371–380 (Springer International Publishing, 2019).
9. Consortium, M. MONAI: Medical Open Network for AI. (2021). doi:10.5281/zenodo.4323058
Figures