2531
SUPER-IVIM-DC, A supervised deep-learning with data consistency approach for IVIM model parameter estimation from Diffusion-Weighted MRI data1Faculty of Biomedical Engineering, Technion-IIT, Haifa, Israel, 2Computational Radiology lab, Boston Children’s Hospital, Harvard Medical School, Boston, MA, United States
Synopsis
Recently, unsupervised deep-learning showed improved performance in estimating the “Intra-Voxel incoherent motion” (IVIM) signal decay model parameters from Diffusion-weighted Magnetic Resonance Imaging (DW-MRI) data compared to classical methods. However, such deep-learning models do not generalize well on acquisitions with a high signal to noise ratio (SNR). In this work, we introduce SUPER-IVIM-DC, a supervised deep learning network coupled with a data consistency term to improve the capacity of deep-learning-based models to generalize the IVIM signal decay model. We demonstrated an improvement in model generalization, accuracy, and homogeneity using simulation, phantom, and in-vivo experiments.
Introduction
Diffusion-weighted Magnetic Resonance Imaging (DW-MRI) is an imaging technique that has the potential to provide important new insights into a multitude of physiological and microstructural properties of the human body [1]. The “Intra-Voxel Incoherent Motion” [2] (IVIM) model relates the observed DW-MRI signal decay to parameters that indirectly reflect blood flow in the capillaries, capillaries volume fraction, and cellular density. These are important biomarkers in several clinical domains such as oncology [3], hepatology [4] and gastroenterology [5], [6]. Least-squares methods for independent voxel-wise fitting of the IVIM model lead to imprecise parameter estimates. While Bayesian approaches demonstrate their potential in producing more accurate estimates, their computational burden is heavy. These challenges diminish the practical usage of IVIM analysis in the clinical environment. Barbieri et al. [7] suggested a deep-neural-network (DNN), IVIM-NET, that demonstrated improved results in terms of better visual assessment, lower root-mean-squared-error (RMSE) for estimation from simulation data with low signal to noise ratio (SNR) and faster computation time in reference to the Bayesian approach. However, it uses an unsupervised-learning approach which may result in suboptimal performance and does not perform as well on high SNR. We present SUPER-IVIM-DC a DNN that integrates both supervised learning and unsupervised-learning approaches for training with a data consistency term aiming to improve DNN capacity to estimate the IVIM parameters from DW-MRI data.Methods
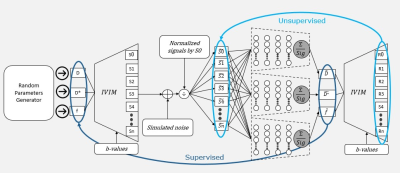
We use the optimized IVIM-NET architecture suggested by Kaandorp et al. [8]. We introduce a supervised loss function with a data consistency term, SUPER-IVIM-DC, computed as the weighted sum of the mean squared error between the ground-truth IVIM parameter values and the IVIM parameter estimations and the mean squared error between the normalized input signal and the reconstructed signal. Alpha coefficients were used to properly scale the contribution of the prediction error of each parameter and the data consistency term as described in the equation:$$\underbrace{\underbrace{\alpha_{D^*}\sum(D^*-\hat{D^*}) + \alpha_{D}\sum(D-\hat{D}) + \alpha_{f}\sum(f-\hat{f})}_{Supervised \ loss} + \alpha_{Recon}\underbrace{\sum(F(\theta)-F(\hat{\theta}))}_{Reconstruction \ loss}}_{SUPER-IVIM-DC \ loss}$$
We followed the IVIM-NET training approach to train SUPER-IVIM-DC. We evaluated the networks using three experiments. First, we tested the noise response of the networks on computer-generated simulation data. We use the same b-values as used in the IVIM-NET for training [8]. For evaluation, 5000 samples were used as an input for the networks. The input data was correlated to the network’s SNR. We used the normalized root-mean-squared error (NRMSE) as the evaluation metric. Second, we tested the bi-exponential fitting performance of the networks using a phantom. The phantom was comprised of 6 small vials of different liquid substances [9], [10]. We scanned the phantom using a 9.4T/20 Animal MRI scanner (Bruker Biospec, Ettlingen, Germany) with the following b-values: {100, 300, 500, 700, 900, 1100, 1300, 1500, 1900, 2100, 2300, 2500} and retrained the networks with this set of b-values. We selected a region-of-interest (ROI) from a relatively homogeneous area in each of the six vials and used it as an input for the DNN. The IVIM parameters were evaluated and assigned in the IVIM equation to create an IVIM curve estimation and coefficient of variance (CV) of every ROI calculated. Last, we tested the networks' homogeneity estimations on 9 clinical DW-MRI studies [11]. The in-vivo abdominal imaging was performed using a 1.5 [T] (Magnetom Avanto, Siemens Medical Solutions, Erlangen, Germany) with the following b-values: {0, 50,100,200, 400,600,800}. A single slice containing the kidneys, spleen, and liver was selected. A homogenous ROI was marked on every organ and used as input. Statistical analysis was performed on every ROI for each IVIM parameter estimation.
Results
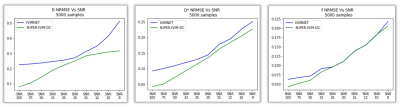
Figure 2 summarizes the results of our simulation studies. Our SUPER-IVIM-DC reduced the median NRMSE compared to IVIM-NET for all IVIM parameters and for all SNR conditions tested in the range of 8-100. Specifically, for SNR 100, SUPER-IVIM-DC reduced the NRMSE in D by 64% (0.08 vs. 0.23, p<1e-9), for D* by 54% (0.04 vs. 0.09 p<1e-9), and for f by 31% (0.04 vs. 0.06 p<1e-9).Our phantom study shows that SUPER-IVIM-DC reduced the CV mean value compared to IVIM-NET for D* and f. Specifically, for the 9% dairy cream vial, SUPER-IVIM-DC reduced the CV mean value in D* by 52% (0.02 vs. 0.04), for f by 92% (0.009 vs. 0.115), and with similar results on D (0.033 vs 0.031).
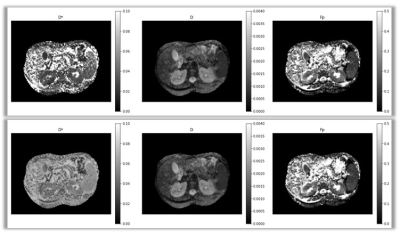
Figure 3 depicts the IVIM parametric maps estimated from clinical abdominal DW-MRI data by SUPER-IVIM-DC (bottom row) and by IVIM-NET (top row). SUPER-IVIM-DC reduced the CV median value compared to IVIM-NET. The major difference was measured on the spleen. SUPER-IVIM-DC reduced the D* CV median value by 73% compared to the IVIM-NET (0.05 vs 0.198 p<1e-4).
Conclusions
SUPER-IVIM-DC has the potential to enable more accurate IVIM parameter estimates compared to IVIM-NET. Our simulation study showed that SUPER-IVIM-DC produces more accurate IVIM parameter estimations even for high SNR data with lower NRMSE on D and D* compared to IVIM-NET. Our phantom study showed that D* and f estimations had a lower CV compared to IVIM-NET and our clinical studies showed lower D* CV estimation on kidneys, spleen, and liver. IVIM maps visual assessment showed the main benefit of SUPER-IVIM-DC, as the D* estimation had the greatest impact on the maps created.Acknowledgements
This research was supported in part by a grant from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel.References
[1] D. M. Koh, D. J. Collins, and M. R. Orton, “Intravoxel incoherent motion in body diffusion-weighted MRI: Reality and challenges,” Am. J. Roentgenol., vol. 196, no. 6, pp. 1351–1361, 2011, doi: 10.2214/AJR.10.5515.
[2] L. M. Le Bihan, Breton E, Lallemand D, Aubin ML, Vignaud J, “Separation of diffusion and perfusion in intravoxel incoherent motion MR imagingSeparation of diffusion and perfusion in intravoxel incoherent motion MR imaging,” in RSNA, 1988, vol. 1, no. 2, pp. 12–17.
[3] D. M. Koh and D. J. Collins, “Diffusion-weighted MRI in the body: Applications and challenges in oncology,” Am. J. Roentgenol., vol. 188, no. 6, pp. 1622–1635, 2007, doi: 10.2214/AJR.06.1403.
[4] J. Patel, E. E. Sigmund, H. Rusinek, M. Oei, J. S. Babb, and B. Taouli, “Diagnosis of cirrhosis with intravoxel incoherent motion diffusion MRI and dynamic contrast-enhanced MRI alone and in combination: Preliminary experience,” J. Magn. Reson. Imaging, vol. 31, no. 3, pp. 589–600, 2010, doi: 10.1002/jmri.22081.
[5] M. Freiman et al., “Characterization of fast and slow diffusion from diffusion-weighted MRI of pediatric Crohn’s disease,” J. Magn. Reson. Imaging, vol. 37, no. 1, pp. 156–163, 2013, doi: 10.1002/jmri.23781.
[6] M. C. Zhang et al., “IVIM with fractional perfusion as a novel biomarker for detecting and grading intestinal fibrosis in Crohn’s disease,” Eur. Radiol., vol. 29, no. 6, pp. 3069–3078, 2019, doi: 10.1007/s00330-018-5848-6.
[7] S. Barbieri, O. J. Gurney-Champion, R. Klaassen, and H. C. Thoeny, “Deep learning how to fit an intravoxel incoherent motion model to diffusion-weighted MRI,” Magn. Reson. Med., vol. 83, no. 1, pp. 312–321, 2020, doi: 10.1002/mrm.27910.
[8] M. P. T. Kaandorp et al., “Improved unsupervised physics-informed deep learning for intravoxel incoherent motion modeling and evaluation in pancreatic cancer patients,” Magn. Reson. Med., vol. 86, no. 4, pp. 2250–2265, 2021, doi: 10.1002/mrm.28852.
[9] Z. Ababneh, M. Haque, S. E. Maier, and R. V. Mulkern, “Dairy cream as a phantom material for biexponential diffusion decay,” Magn. Reson. Mater. Physics, Biol. Med., vol. 17, no. 2, pp. 95–100, 2004, doi: 10.1007/s10334-004-0063-7.
[10] G. S. Ioannidis, K. Nikiforaki, G. Kalaitzakis, A. Karantanas, K. Marias, and T. G. Maris, “Inverse Laplace transform and multiexponential fitting analysis of T2 relaxometry data: a phantom study with aqueous and fat containing samples,” Eur. Radiol. Exp., vol. 4, no. 1, 2020, doi: 10.1186/s41747-020-00154-5.
[11] V. Taimouri et al., “Spatially constrained incoherent motion method improves diffusion-weighted MRI signal decay analysis in the liver and spleen,” Med. Phys., vol. 42, no. 4, pp. 1895–1903, 2015, doi: 10.1118/1.4915495.
Figures