2529
DCE-DRONE: Perfusion MRI Parameter Estimation using a DRONE Neural Network
Soudabeh Kargar1, Ouri Cohen2, and Ricardo Otazo2
1MSKCC, NEW YORK, NY, United States, 2MSKCC, New York, NY, United States
1MSKCC, NEW YORK, NY, United States, 2MSKCC, New York, NY, United States
Synopsis
In this work, we demonstrate the estimation of DCE acquired perfusion parameters using a DRONE neural network trained on numeric simulated data. Experiments in a digital phantom are used to demonstrate the feasibility of our approach and the clinical utility is shown in subjects with gynecological tumors.
Introduction
Dynamic Contrast Enhanced (DCE) MRI is an important component of multi-parametric MRI for cancer diagnosis and assessment of treatment response [1]. DCE-MRI is usually performed with a spoiled gradient echo sequence and the temporal signal in each pixel is converted to contrast agent concentration for perfusion analysis using signal model. Accurate conversion requires a dynamic or pre-contrast T1 map. In practice, a constant T1 is often selected for the blood and the tissue of interest under the assumption of linearity between signal and contrast concentration due to low contrast concentration. In cases where this assumption fails, errors are introduced into the quantification. Here we propose to use the DRONE [2] neural network (NN) to estimate the perfusion parameters from the DCE time-series while accounting for tissue-specific T1 and steady-state magnetization S0.Methods
A four-layer DRONE network with 300 nodes per layer was defined in Pytorch and trained to convergence using the ADAM optimizer [3]. A training dictionary with 300,000 entries was defined with parameters selected from the following ranges: Ktrans: [0.15,1], vp : [0.001,0.2], ve: [0.05,0.6], T1 : [30,1250], S0 : [0.01,1]. The parameters were used as input to the extended Tofts model [4] with the Parker population-based Arterial Input Function (AIF) [5] to generate the synthetic concentration curves C(t) which were then converted to a magnetization signal using the SPGR signal equation (Eq.1,2). The network was trained for 9000 epochs. Zero mean Gaussian noise was added to the training to promote robust learning. A flowchart diagram of the dictionary generation, training, and testing is shown in Fig. 1.$$ S(t) = S_0 \frac{sin(\alpha)(1-\exp^{-TR(\frac{1}{T_{10}}+r_1C(t))})}{1-cos(\alpha)\exp^{-TR(\frac{1}{T_{10}}+r_1C(t))}} \space \space \space \space Eq.1 $$
$$ \frac{1}{T_1} = \frac{1}{T_{10}}+r_1C(t) \space \space \space \space Eq.2 $$
The DRONE input layer was tailored to the number of time-points in the measured DCE-MRI data. The network’s output consisted of the Ktrans, vp, ve parameters. The schematic of the NN is shown in Fig.2. To test the accuracy of the DRONE reconstruction, a Brainweb-based [6] digital brain phantom was modified to include a variety of perfusion parameters values (Fig.3).Two female subjects with cervical cancer were recruited for this study and gave informed consent in accordance with the institutional IRB. The subjects were scanned on a 3T GE Premier scanner (GE Healthcare, Milwaukee, WI) with the radial stack of stars sequence and the raw data reconstructed with the Golden-angle Radial Sparse Parallel (GRASP) [7] method with temporal resolution of seconds (60 time frames). The Repetition Time (TR) was set to 3.03 ms, and the flip angle (α) to 9°. The network was trained on the simulated data with the same temporal resolution and using physiologically relevant perfusion and T1 values. A contrast agent relativity of r1 = 4.0mM-1s-1 was used for the signal to contrast conversion.
Results
The estimated maps in the digital phantom in comparison to the reference values are shown in Fig.3 along with the estimation error calculated as (true-estimated)/true x 100. The correlation between the estimated and reference values was also measured. Estimated perfusion parameters in the cancer subjects are shown in Fig.4 and Fig.5. The T1 and S0 results of the in vivo results were noisy and needs further fine tuning of the network. Adding the flip angle as part of the training could improve the T1/S0 discrimination.Discussion
Estimation of perfusion parameters using DRONE offers improved accuracy and robustness to noise with minimal processing time. The use of simulated training data allows inclusion of, potentially confounding, instrumental parameters (e.g. B1) in the training to mitigate its effect. Fine tuning the network parameters (training dictionary size, perfusion parameters range, etc.) may improve the results further. Similarly, although in this work we used the Parker population AIF for simplicity, the AIF could also be incorporated into the network training to enable estimation of a patient-specific AIF. This will be explored in future work.Conclusion
This is an initial proof-of-concept for estimation of DCE perfusion parameters using a model-trained deep neural network. Incorporating additional parameters into the training is expected to improve the accuracy and reproducibility of the estimated perfusion parameters.Acknowledgements
Kargar S. and Cohen O. contributed equally to this work.References
- Leach MO, Brindle KM, Evelhoch JL, Griffiths JR, Horsman MR, Jackson A, Jayson GC, Judson IR, Knopp MV, Maxwell RJ, McIntyre D, Padhani AR, Price P, Rathbone R, Rustin GJ, Tofts PS, Tozer GM, Vennart W, Waterton JC, Williams SR, Workman P; Pharmacodynamic/Pharmacokinetic Technologies Advisory Committee, Drug Development Office, Cancer Research UK. The assessment of antiangiogenic and antivascular therapies in early-stage clinical trials using magnetic resonance imaging: issues and recommendations. Br J Cancer. 2005 May 9;92(9):1599-610. doi: 10.1038/sj.bjc.6602550.
- Kingma DP,BaJL. Adam: A method for stochastic optimization; 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings 2015
- Cohen O, Zhu B, Rosen MS. "MR fingerprinting Deep RecOnstruction NEtwork (DRONE)". Magn Reson Med. 2018 Sep;80(3):885-894. doi: 10.1002/mrm.27198.
- Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997 Jan-Feb;7(1):91-101. doi: 10.1002/jmri.1880070113.
- Parker GJ, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med. 2006 Nov;56(5):993-1000. doi: 10.1002/mrm.21066.
- https://brainweb.bic.mni.mcgill.ca/brainweb/
- Feng L, Grimm R, Block KT, Chandarana H, Kim S, Xu J, Axel L, Sodickson DK, Otazo R. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magnetic Resonance in Medicine, (2014), 72(3). doi: 10.1002/mrm.24980
Figures
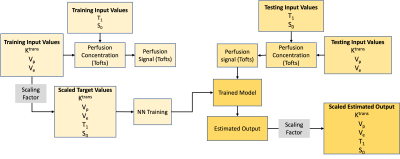
Fig 1. The flowchart of the dictionary generation, training, and testing with neural network.
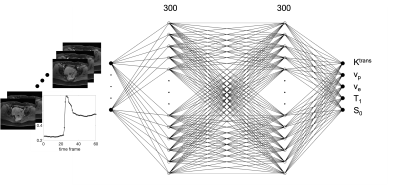
Fig. 2 The fully connected neural network with 60 nodes for input for a signal with 60 time frames, two 300 node hidden layers, and 5 output nodes for Ktrans, vp, ve, T1, and S0.
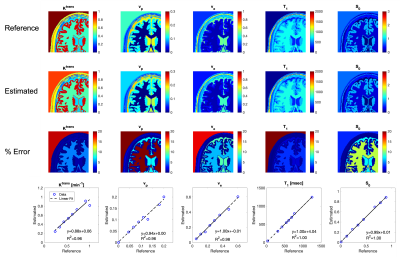
Fig. 3 Numeric simulation results. The reference values, estimated with Deep Learning Method, and the error percentage of the maps are shown. The linear regression of the estimated vs. reference values are shown on the bottom row.

Fig. 4 . The in vivo results for the deep learning method shown for a cervical cancer tumor in five consecutive slices.
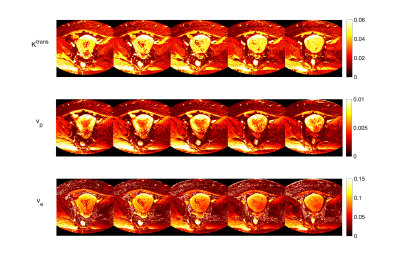
Fig. 5 . The in vivo results for the deep learning method shown for a cervical cancer tumor in five consecutive slices.
DOI: https://doi.org/10.58530/2022/2529