2525
Fully Connected Deep Neural Network combined with Segmented Least Square Fitting for improved extraction of IVIM Parameters1Istituto di Tecnologie Biomediche, Consiglio Nazionale delle Ricerche, Segrate, Italy, 2Radiology, Northwestern University, Chicago, IL, United States
Synopsis
Voxel-by-voxel fitting of intravoxel incoherent motion (IVIM) MRI data using a bi-exponential model is challenging especially with low signal-to-noise ratio (SNR) diffusion-MR images. We propose to combine and use a supervised Deep Neural Network (DNN) approach to increase SNR of the acquired images and thus improve extraction of reliable parameter estimation using a segmented least square fitting algorithm. The effectiveness of the proposed method was demonstrated in both simulated and acquired in vivo data. The proposed approach is promising and can increase performance of the fitting algorithms especially for the case of images with high background noise.
Introduction
IVIM model uses a bi-exponential diffusion MR signal representation to account for diffusion and perfusion-related tissue properties1. While the IVIM technique has been used for several biomedical applications the bi-exponential fitting process can be challenging and the estimation of perfusion related parameters is not always reliable due to the high background noise. “Segmented” least square fitting techniques, which consider the diffusion signal at high and low b-values separately, have been proposed as a valuable alternative for IVIM parameters estimation2. Nevertheless, the segmented approach still underperforms especially when working with low SNR images. The purpose of this work is to demonstrate the potential of a supervised, trained and fully connected DNN to improve the SNR of original data sets and to increase overall reliability of the segmented fitting approach.Methods
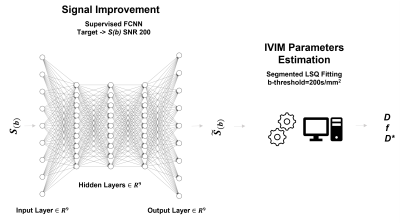
Deep Neural NetworkA three hidden layer feed-forward neural network with 9 nodes for each layer having a sigmoid activation function for the hidden layers and a linear function for the output layer was implemented in MATLAB. The training was performed using Levenberg-Marquardt backpropagation algorithm and mean square root error as loss function. The network was trained using 5000 simulated images (1000 for each SNR ranging from 10 to 150), splitting the data 70/15/15% for training/validation/test. Images at SNR 200 were considered as the final target. The proposed neural network architecture is displayed in figure 1.
Simulations
Numerical phantoms were generated using a MatLab R2020a custom made script. Starting from physiologically typical mammal brain D values in the range [0.0005-0.002 mm2/s], D*[0.005-0.1] and f [0.025-0.4], diffusion weighted images were computed for different b-values (0, 25, 50, 75, 100, 150,300, 800, 1000 s/mm2). Rician noise was added to create the signal diffusion weighted images with different SNRs (10, 25, 50, 100, 150, 200). The Shepp-Logan phantom was used in our simulations; each region of interest was characterized by a randomly generated parameters triplet.
In vivo Data
Experimental procedures involving animals complied with Northwestern’s IACUC guidelines. MRI acquisitions were performed on a 7T ClinScan MRI scanner (Bruker, Germany) equipped with a 12 cm diameter gradient coil system (max strength 115 mT/m) using a four-channel phase-array receiver coil. A volume quadrature coil was used for transmission. The imaging protocol included a multiple b-values (bs =0, 25, 50, 75, 100, 150, 300, 800, 1000) SE-EPI diffusion weighted images (TR/TE=3500/27 ms, flip-angle=90, averages=4, slice-thickness=1 mm, voxel-size=0.282x0.282 mm2).
Segmented Fitting
The least square segmented method implementation by Jalnefjord 3 using custom made MATLAB functions was used to fit the bi-exponential decay of the diffusion MR images. In the first step, data from b values below 200 s/mm2 were omitted to estimate the parameter D. In the second step, f and D* were jointly estimated using the whole signal decay.
Performance Assessment
The DNN performances were assessed on test images using as accuracy metrics the Root Mean Square Error (RMSE), the SNR and the peak SNR (pSNR). To evaluate the fitting performances, the Median Error (ME), the Coefficient of Variation (CV) and the Maximum Absolute Error (MAE) were calculated. Ground truth parameters were considered as reference. Quantitative analysis was implemented in MATLAB.
Results
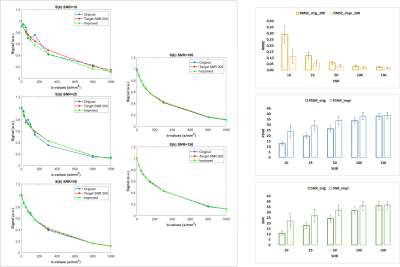
DNN PerformanceAs shown in figure 2, the use of the proposed DNN increases the quality of the diffusion signal especially at low SNRs. RMSE increases up to more than 100% and its increment is remarkable especially at low SNRs. Analogously, SNR and pSNR increases up to 80% at low SNRs. Whereas, as expected, these differences are smaller at high SNRs.
Fitting Accuracy
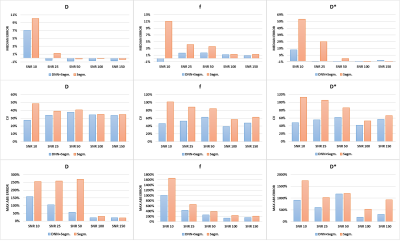
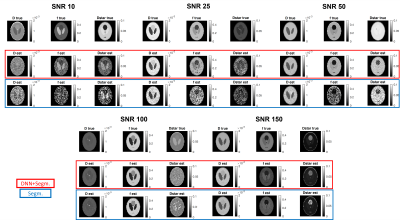
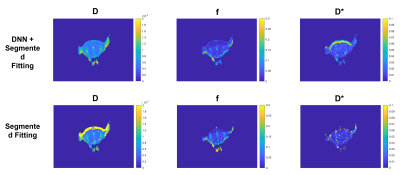
The accuracy of the proposed method was evaluated quantitatively using simulated data. As shown in figure 3, the use of the DNN reduces the CVs and MAEs in every condition and this is more evident at low SNRs. Considering the ME, in most of the conditions its value is decreased and this result is much more evident in the case of f and D* estimation at low SNRs. Figure 4 shows some representative examples comparing the quality of the parameter maps generated using the proposed DNN approach with those obtained using only the standard segmented fitting algorithm.
In vivo data
As shown in figure 5, the proposed approach also improves the overall quality of the IVIM parametric maps generated from in vivo mouse brain MRI acquisitions.
Discussion
The proposed DNN-based approach combined with segmented least square fitting can improve the reliability of the IVIM parameters estimation by improving the overall image SNR and thus increasing the overall fitting performance of the segmented least square algorithm. Importantly the described voxel-by-voxel DNN approach reduces background noise without introducing significant blurring effects in the images and therefore preserving biologically relevant features. The method is easy to implement and, following DNN training, can be deployed for image analysis with relatively fast computational times. Changes in experimental parameters such as choice of b-values require re-training of DNN.Conclusion
The proposed combination of DNN with a segmented fitting methods approach was demonstrated to be effective to increase the robustness of IVIM results when compared to those obtained using only standard fitting algorithms. This is particularly useful when processing MR images with higher background noise typically obtained in vivo.Acknowledgements
No acknowledgement found.References
1. Le Bihan, D. (2019). What can we see with IVIM MRI?. Neuroimage, 187, 56-67.
2. Barbieri, S., Donati, O. F., Froehlich, J. M., & Thoeny, H. C. (2016). Impact of the calculation algorithm on biexponential fitting of diffusion‐weighted MRI in upper abdominal organs. Magnetic resonance in medicine, 75(5), 2175-2184.
3. Jalnefjord, O., Andersson, M., Montelius, M., Starck, G., Elf, A. K., Johanson, V., ... & Ljungberg, M. (2018). Comparison of methods for estimation of the intravoxel incoherent motion (IVIM) diffusion coefficient (D) and perfusion fraction (f). Magnetic Resonance Materials in Physics, Biology and Medicine, 31(6), 715-723.
Figures