2501
A multi-task generative network for simultaneous post-contrast MR image synthesis and tumor segmentation: application to brainstem glioma1MR Clinical Science, Philips Healthcare, Suzhou, China, 2Department of Radiology, Beijing Tiantan Hospital, Capital Medical University, Beijing, China
Synopsis
To reduce the exposure of Gadolinium-based Contrast Agents (GBCAs) in brainstem glioma detection and provide high-resolution contrast information, we propose a novel multi-task generative network for contrast-enhanced T1-weight MR synthesis on brainstem glioma images. The proposed network can simultaneously synthesize the high-resolution contrast-enhanced image and the segmentation mask of brainstem glioma lesions.
Introduction
Gadolinium-based contrast agents (GBCAs) have been widely used for brain MR image enhancement. However, the usage of GBCAs has raised the safety concerns. Since advanced renal disease and the development of nephrogenic systemic fibrosis (NFS) are related to high exposure to GBCAs1, some studies suggested that GBCA dose should be minimized 2. In this study, we propose a multi-task method for contrast enhanced T1-weight synthesis based on pre-contrast T1-weight (T1w), T2 weight (T2w) and Arterial Spin Labeling (ASL) images.Methods
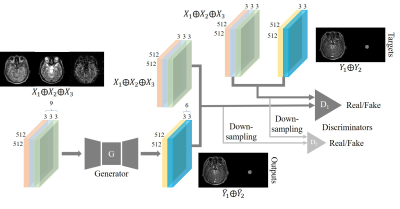
A cohort of 30 brainstem glioma (BSG) patients were included in this study. Consent forms were obtained from all patients. The whole brain MR scans were acquired on multiple imaging platforms including Philips, GE and Siemens. A total of 720 axial image sets with multi-contrast were included in this study. 24 patients’ data were randomly selected for training and the rest for testing. We employed two experienced radiologists to annotate the segmentation mask of BSG lesion. Pre-contrast T1w, T2w and ASL were used as input images. The post-contrast T1w and corresponding manual lesion mask were used as target images. The images were firstly resized to 512 x 512 matrix, and Elastix tool3 was used to register the multi-contrast images within each patient. The training images were augmented with random flip. We proposed a multi-task conditional generative adversarial network (mt-cGAN) as shown in Fig.1. It contained a generator G4 and two multi-scale discriminators D={D1, D2} which were based on patchGANs. D1 and D2 with the target Y with high resolution 512 x 512 and low resolution 256 x 256, respectively. The overall loss function was defined as $$$ L=\sum_{k=1,2}L_{cGAN}(G,D_{k})+\lambda_{FM}\sum_{k=1,2}L_{FM}(G,D_{k})+\lambda_{PER}L_{PER}(G,V)$$$, where $$$L_{FM}$$$ denotes the feature matching loss, $$$L_{PER}$$$ denotes the perceptual loss, V is the pretrained VGG-19 network5, and $$$ \lambda_{FM}=10$$$, $$$ \lambda_{PER}=10$$$ as empirical setting. Two metrics were employed to measure the distance between the synthesized images and the ground truth images: structural similarity index measurement (SSIM) and peak signal noise ratio (PSNR). Dice overlap was used to evaluate the automated segmentation on BSG lesions.Results
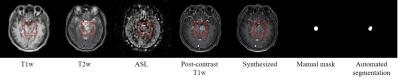
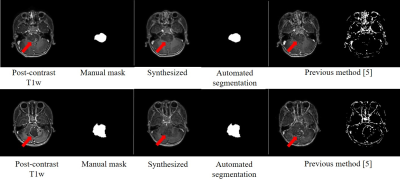
Fig. 2 shows an example of the synthesized image with fine structures and post-contrast like information at the BSG lesion; meanwhile, the BSG automated segmentation matched the manual mask well. Fig. 3 shows typical cases from the testing set. Compared with the result from previous method5, our proposed network can synthesize more realistic contrast-enhanced like T1-weight images with BSG lesions. For the testing set, the synthesized images had PSNR of $$$26.30±2.31$$$ and SSIM of $$$0.86±0.04$$$, which outperformed than the results from previously published method5 (in which, PSNR was $$$24.24±2.07$$$ and SSIM was $$$0.81±0.05$$$). The Dice coefficient of the lesion segmentation reached $$$0.83 ±0.33$$$.Discussion and Conclusion
In this study, we proposed a multi-task generative network to predict contrast-enhanced T1-weight image from multi-contrast MR images, and segmented the region of brainstem glioma simultaneously. The evaluation metrics demonstrate that our proposed network can synthesize the post-contrast T1w image with high quality and segment the lesion with satisfied Dice score. It is potential as a screening step to predict contrast enhanced information as well as to automatically segment the BSG lesion before the patients have to perform an injection for contrast-enhanced MR examination. Further investigation will focus on the validation of broader BSG dataset and to explore the connection between the BSG lesion and the related gene expressions.Acknowledgements
No.References
1. Fraum, T.J., et al., Gadolinium‐based contrast agents: a comprehensive risk assessment. 2017. 46(2): p. 338-353.
2. Gong, E., et al., Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reson Imaging, 2018. 48(2): p. 330-340.
3. Klein, S., et al., Elastix: a toolbox for intensity-based medical image registration. 2009. 29(1): p. 196-205.
4. Zhu, J.-Y., et al. Unpaired image-to-image translation using cycle-consistent adversarial networks. in Proceedings of the IEEE international conference on computer vision. 2017.
5. Chen, C., et al., Synthesizing MR Image Contrast Enhancement Using 3D High-resolution ConvNets. 2021.
Figures