2468
Quiet quantitative susceptibility mapping with Looping Star
Nikou Louise Damestani1, Ana Beatriz Solana2, Florian Wiesinger2, Brice Fernandez3, Steven Charles Rees Williams1, and David John Lythgoe1
1Department of Neuroimaging, King's College London, London, United Kingdom, 2GE Healthcare, Munich, Germany, 3GE Healthcare, Paris, France
1Department of Neuroimaging, King's College London, London, United Kingdom, 2GE Healthcare, Munich, Germany, 3GE Healthcare, Paris, France
Synopsis
Looping Star is an acoustically quiet T2*-weighted acquisition technique, which has primarily been used to demonstrate functional sensitivity. We present the application of Looping Star to quantitative susceptibility mapping in a small cohort, directly comparing its outcomes with a conventional susceptibility-weighted imaging technique. We found that Looping Star produced comparable quantitative values in subcortical regions in comparison with conventional susceptibility-weighted imaging.
Introduction
Looping Star is a novel, acoustically quiet, T2*-weighted pulse sequence that has not only successfully demonstrated functional sensitivity1-3, but also preliminary indications towards use in quantitative susceptibility mapping1. Quantitative susceptibility mapping (QSM) is a clinically useful tool across numerous patient cohorts, particularly for the identification of iron deposition in the brain that could be linked to distinct disorders. However, conventional susceptibility-weighted imaging is incredibly loud, potentially leading to poor image quality due to discomfort-induced motion.The extension of Looping Star to quantitative susceptibility mapping would greatly benefit studies including patient cohorts who are sound averse, given its quiet nature, although a direct comparison with a conventional susceptibility-weighted imaging method has not yet been performed. This work presents the first direct comparison of the most recent iteration of the Looping Star pulse sequence, known as coherence-resolved Looping Star (CRLS)4, with conventional gradient echo Susceptibility-Weighted ANgiography (SWAN) in a small cohort.
Methods
Five participants volunteered for this study and were scanned using a 12-channel receive-only head coil and 3T MR750 GE Healthcare scanner (GE Healthcare, Milwaukee, WI). One participant was excluded from analysis due to artefacts in both acquisitions.CRLS acquisition parameters included: 2 spokes per loop, FID + 9 echoes, ± 46.875 kHz readout bandwidth, 1 mm isotropic resolution, FOV = 19.2 cm, matrix size = 192 x 192, ΔTE (ms) = 4.8 ms, FA = 3°, scan time 10 mins 30 s. The SWAN acquisition parameters were: 256 x 256 matrix size, ± 62.5 kHz readout bandwidth, 138 slices, 13 echoes, FA = 20°, ΔTE (ms) = 3.3 ms, FOV = 25.6 cm, in-plane voxel size = 1 mm, scan time 9 mins 58 s.
Nearest-neighbour gridding reconstruction was employed for CRLS, including coil combination using a simple sum of squares technique with the sensitivity maps calculated from a calibration region in the centre of k-space5. The SWAN data were reconstructed using standard GE Healthcare online reconstruction. Acoustic noise was measured at the centre of the scanner bore using an MR-compatible sound level meter (Casella Solutions, Bedford, UK).
Both datasets were processed using a pipeline in SuscEptibility mapping PIpeline tool for phase images (SEPIA)6, including:
- Echo phase combination using optimum weights from SNR weighting.
- Laplacian phase unwrapping using MEDI7.
- Background field removal with VSHARP via STI suite8, with spherical mean value (SMV) size 5mm
- Removal of the residual using a 3D polynomial of order 4.
- QSM using MEDI7 with morphology-enabled dipole inversion regularisation parameter value 1000 and 5 x 5 x 5 voxel zero-padding. CSF was used as the reference tissue. .
Brain masking was performed using the FSL Brain Extraction Tool9. The R2* map was computed using MEDI, which uses auto-regression on linear operations mono-exponential decay fitting function (ARLO)10.
Average and standard deviation values for subcortical regions were produced by normalising the QSM and R2* maps to MNI space for each participant using SPM-12, after co-registration with each T1-weighted anatomical image, followed by averaging the values across the anatomical regions of interest. The regions of interest were 5 mm radius spheres centred in the subcortical structures, defined by the Harvard-Oxford subcortical atlas. The regions selected where the putamen, thalamus and caudate nucleus.
Results
The acoustic noise measures (LAeq) for CRLS QSM were 81.9 dB(A) and for SWAN were 110.4 dB(A), compared with the ambient scan room noise at 65 dB(A). Figure 1 shows an example of a single subject magnitude and phase image for a single echo, as well as the QSM and R2* map. Comparable contrast was identified between the modalities. Figure 2 illustrates the same comparison, though focused on subcortical structures given their differing susceptibility relative to cortical tissues11-12.Figure 3 highlights the average and standard deviation R2* values across participants for each modality. Good agreement can be seen between techniques, and the quantitative values identified were consistent with literature13. We found high variability in our susceptibility measures across participants, indicating that a unique pipeline may be required for Looping Star.
Conclusions
This study demonstrates the capability of multi-echo coherence-resolved Looping Star for T2*-based imaging beyond functional MRI. Future work should focus on optimisation of the pre-processing pipeline for Looping Star and methods for image quality improvement, as well as extension to larger cohorts.Acknowledgements
This presentation represents independent research supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. Nikou is in receipt of a PhD studentship funded by the NIHR Maudsley Biomedical Research Centre.The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. Florian Wiesinger, Ana Beatriz Solana and Brice Fernandez are employed by GE Healthcare.References
- Wiesinger, F., Menini, A. and Solana, A.B., 2019. Looping star. Magnetic resonance in medicine, 81(1), pp.57-68.
- Damestani, N.L., O'Daly, O., Solana, A.B., Wiesinger, F., Lythgoe, D.J., Hill, S., de Lara Rubio, A., Makovac, E., Williams, S.C. and Zelaya, F., 2021. Revealing the mechanisms behind novel auditory stimuli discrimination: An evaluation of silent functional MRI using looping star. Human Brain Mapping, 42(9), pp.2833-2850.
- Dionisio‐Parra, B., Wiesinger, F., Sämann, P.G., Czisch, M. and Solana, A.B., 2020. Looping star fMRI in cognitive tasks and resting state. Journal of Magnetic Resonance Imaging, 52(3), pp.739-751.
- Damestani, N.L., Lythgoe, D.J., Solana, A.B., Fernandez, B., Williams, S.C.R., Zelaya, F., Wiesinger, F. 2021. Coherence-resolved Looping Star (CRLS): Improvements to image quality for silent structural and functional neuroimaging. In Proceedings of ISMRM Virtual Meeting 2021. Poster 2677.
- McKenzie, C.A., Yeh, E.N., Ohliger, M.A., Price, M.D. and Sodickson, D.K., 2002. Self‐calibrating parallel imaging with automatic coil sensitivity extraction. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 47(3), pp.529-538.
- Chan, K.S. and Marques, J.P., 2021. SEPIA—susceptibility mapping pipeline tool for phase images. Neuroimage, 227, p.117611.
- Liu, T., Liu, J., De Rochefort, L., Spincemaille, P., Khalidov, I., Ledoux, J.R. and Wang, Y., 2011. Morphology enabled dipole inversion (MEDI) from a single‐angle acquisition: comparison with COSMOS in human brain imaging. Magnetic resonance in medicine, 66(3), pp.777-783.
- Li, W., Avram, A.V., Wu, B., Xiao, X. and Liu, C., 2014. Integrated Laplacian‐based phase unwrapping and background phase removal for quantitative susceptibility mapping. NMR in Biomedicine, 27(2), pp.219-227.
- Jenkinson, M., Beckmann, C.F., Behrens, T.E., Woolrich, M.W. and Smith, S.M., 2012. Fsl. Neuroimage, 62(2), pp.782-790.
- Pei, M., Nguyen, T.D., Thimmappa, N.D., Salustri, C., Dong, F., Cooper, M.A., Li, J., Prince, M.R. and Wang, Y., 2015. Algorithm for fast monoexponential fitting based on auto‐regression on linear operations (ARLO) of data. Magnetic resonance in medicine, 73(2), pp.843-850.
- Cohen-Adad, J., 2014. What can we learn from T2* maps of the cortex?. Neuroimage, 93, pp.189-200.
- Feng, X., Deistung, A. and Reichenbach, J.R., 2018. Quantitative susceptibility mapping (QSM) and R2* in the human brain at 3 T: Evaluation of intra-scanner repeatability. Zeitschrift für Medizinische Physik, 28(1), pp.36-48.
- Hagemeier, J., Zivadinov, R., Dwyer, M.G., Polak, P., Bergsland, N., Weinstock-Guttman, B., Zalis, J., Deistung, A., Reichenbach, J.R. and Schweser, F., 2018. Changes of deep gray matter magnetic susceptibility over 2 years in multiple sclerosis and healthy control brain. NeuroImage: Clinical, 18, pp.1007-1016.
Figures
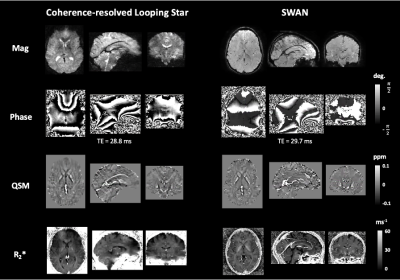
Figure 1: Example of single subject magnitude, phase, quantitative susceptibility map (QSM) and R2* map of whole brain for each modality. ppm = parts per million, TE = echo time, deg = degrees.
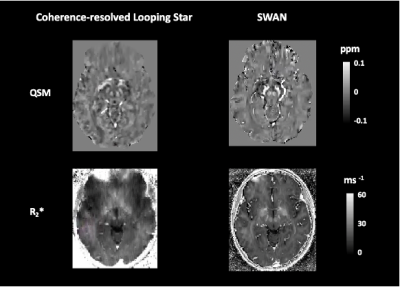
Figure 2: Example of single subject quantitative susceptibility map (QSM) and R2* map of subcortical regions. ppm = parts per million.
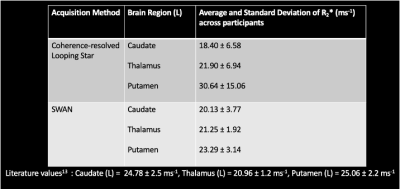
Figure 3: Table of average and standard deviation R2* values for different subcortical regions in the left hemisphere across participants.
DOI: https://doi.org/10.58530/2022/2468