2440
Rapid high-resolution cranial bone MRI using deep-learning prior image reconstruction1Washington University in Saint Louis, Saint Louis, MO, United States
Synopsis
A high-resolution (HR) MRI capable of resolving the detail of bony structures at sub-millimeter resolution is desired. A short MR acquisition results in under-sampled k-space data below the Nyquist rate, leading to artifacts and high noise. We developed an HR reconstruction method regularized by a complex deep-learning prior (RECD). We achieved high-resolution MR (0.6x0.6x0.8mm3) with a one-minute acquisition time. Using images reconstructed from a 5-minute MR scan as the gold standard, we compared the peak signal to noise ratio (PSNR) and similarity index (SSIM) for 1-min RECD and 1-min compressed sensing (CS) reconstructed images. RECD outperformed CS.
Introduction
3D high-resolution cranial bone MRI is a safe alternative to CT for pediatric patients with cranial anomalies without ionizing radiation 1-5. CT imaging only takes seconds or tens of seconds to acquire, while high-resolution MRI capable of resolving details at sub-millimeter resolution usually takes several minutes to cover the whole head. A rapid cranial bone MRI may greatly facilitate a clinical adoption in craniosynostosis and head trauma diagnosis. Accelerated MR acquisition usually samples k-space below the Nyquist rate, leading to artifacts and high noise. To overcome these challenges, compressed sensing (CS) reconstruction methods may be employed 6-8. However, CS becomes less effective when data is severely under-sampled. More recently, deep learning (DL) has been developed for MR image reconstruction 9-16. In this study, we aimed to develop an MR reconstruction regularized by 3D complex image deep learning prior (RECD) to shorten the MRI acquisition time from 5-minutes to 1-minute. The proposed method was evaluated on its ability to detect suture and skull fracture in pediatric patients with craniosynostosis or head trauma.Method
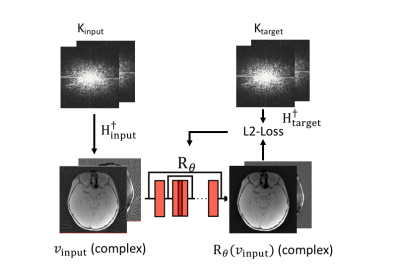
St. Louis Children's hospital pediatric patients (ages three days to 17 years old) diagnosed with head trauma or craniosynostosis were recruited for this study. MRI scans were obtained from 40 pediatric subjects (15 Female). Participants had a Golden Angle (GA) stack-of-stars scan with the following imaging parameters TE/TR = 2.47ms/4.84ms, Flip angle = 3°, BW = 410 Hz/pixel, FOV = 192 mm, 224 slices, voxel size 0.6x0.6x0.8 mm, Number of radial spokes = 400, Total acquisition time = 5:04. A multi-coil non-uniform fast Fourier transform (MCNUFFT) was performed to reconstruct images using 80, 160, 240, 320 and 400 spokes.The RECD method reconstructed images by iteratively minimizing a loss function in Eq. 1.u = argminu ∑i ||FCiu-Ki ||2+ λ L2(Rθ(u), u) (1)
where u is the image to-be-reconstructed, Ci is the coil sensitivity for each coil i, F is the forward encoding operator with the k-space sampling function (Hi=FCi) , Ki is the acquired k-space data by coil i. Rθ is a pre-trained 3D complex image deep learning network, L2 represents L2 loss, and λ is the regularization coefficient. A ResUNet structure, consisting of seven layers with both contraction and expanding paths, was used in Rθ. Rθ has dual-channel inputs for real and imaginary parts of the complex images. The complex images were used because noise has a zero-mean Gaussian distribution. During the training, the k-space data of a 5-minute scan can be divided into disjoint subsets, the first 1-minute as Kinput and the remaining as Ktarget (Fig 1). A forward Fourier transform (MCNUFFT) was used to transform Kinput and Ktarget into complex image vinput and vtarget. Training inputs included 1, 2, 3, and 4-minute MCNUFFT images, and training targets included 4 and 5-minute MCNUFFT images. Rθ was trained using a L2 loss function as described in Eq. 2.
argminθ∑ij L2(Rθ(vinput), vtarget) (2)
Over 200 pairs of different inputs and targets from 28 subjects were used for training . The trained Rθ was then used as a prior to regularize the image reconstruction in an iterative optimization to obtain image u as described in Eq (1). RECD was used to reconstruct the first 80 spokes (1-minute) k-space data for the remaining 12 testing subjects. In addition, a CS reconstruction regularized by a total generalized variation (TGV) prior 8 was applied to the same data. The 400 spokes (5-minutes) MCNUFFT images were used as the gold standard. SSIM and PSNR were used to compare the quality of the CS and RECD reconstructed images using 1-minute data.
Results
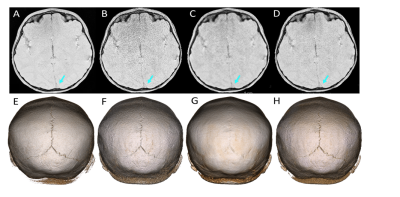
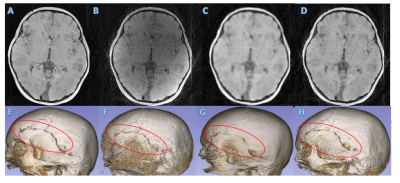
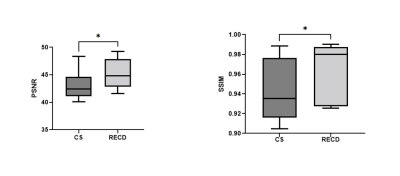
Volumetric and 3D rendered cranial images from a 4.2-year-old female patient with lambdoid sutures (Fig 2) and from a 5-year-old male trauma patient with a fracture (Fig 3) were created. The 3D rendered images were created from inverted MR images using 3D Slicer (5). Compared to the 5-min MCNUFFT images (Fig 2A and 2E, 3A and 3E), the 1-min MCNUFFT had high noise and streaking artifacts (Fig 2B and 2F, 3B and 3F). The 1-min CS reduced noise and artifacts at the expense of image sharpness, resulting in less visible sutures and fractures (Fig 2C and 2G, 3C and 3G). The RECD reconstruction was effective in reducing artifacts and noise while preserving fine structure details (Fig 2D and 2H,3D and 3H). The SSIM and PSNR for CS and RECD were summarized in a box plot in Figure 4. The RECD images have significantly higher PSNR (P=0.012) and SSIM (P=0.024) than the CS.Discussion and Conclusions
We developed an MRI reconstruction method, RECD, to accelerate MR acquisition. The 5-min (400 spokes) and 1 min (80 spokes) scans are at 80% and 16% of Nyquist rates, respectively. The RECD one-minute images closely resembled the 5-minute MRI images in terms of image quality. Our results showed that the proposed RECD approach outperformed a CS approach. We demonstrated that RECD reconstructed one-minute MR images can be used to identify skull fractures and sutures for clinical diagnostic evaluation for craniosynostosis and trauma patients.Acknowledgements
No acknowledgement found.References
1. Kralik SF, Supakul N, Wu IC, Delso G, Radhakrishnan R, Ho CY, Eley KA. Black bone MRI with 3D reconstruction for the detection of skull fractures in children with suspected abusive head trauma. Neuroradiology. 2019;61(1):81-7. Epub 2018/11/09. doi: 10.1007/s00234-018-2127-9. PubMed PMID: 30406272.
2. Dremmen MHG, Wagner MW, Bosemani T, Tekes A, Agostino D, Day E, Soares BP, Huisman T. Does the Addition of a "Black Bone" Sequence to a Fast Multisequence Trauma MR Protocol Allow MRI to Replace CT after Traumatic Brain Injury in Children? AJNR Am J Neuroradiol. 2017;38(11):2187-92. Epub 2017/10/04. doi: 10.3174/ajnr.A5405. PubMed PMID: 28970241.
3. Eley KA, McIntyre AG, Watt-Smith SR, Golding SJ. "Black bone" MRI: a partial flip angle technique for radiation reduction in craniofacial imaging. Br J Radiol. 2012;85(1011):272-8. Epub 2012/03/07. doi: 10.1259/bjr/95110289. PubMed PMID: 22391497; PMCID: PMC3473988.
4. Eley KA, Watt-Smith SR, Sheerin F, Golding SJ. "Black Bone" MRI: a potential alternative to CT with three-dimensional reconstruction of the craniofacial skeleton in the diagnosis of craniosynostosis. Eur Radiol. 2014;24(10):2417-26. Epub 2014/07/21. doi: 10.1007/s00330-014-3286-7. PubMed PMID: 25038852
5. Patel KB, Eldeniz C, Skolnick GB, Jammalamadaka U, Commean PK, Goyal MS, Smyth MD, An H. 3D pediatric cranial bone imaging using high-resolution MRI for visualizing cranial sutures: a pilot study. J Neurosurg Pediatr. 2020 Jun 12:1-7. doi: 10.3171/2020.4.PEDS20131. Epub ahead of print. PMID: 32534502; PMCID: PMC7736460.
6. Donoho DL. Compressed sensing. IEEE Transactions on Information Theory. 2006;52(4):1289-306. doi: 10.1109/TIT.2006.871582.
7. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic resonance in medicine. 2007;58(6):1182-95.
8. Knoll F, Bredies K, Pock T, Stollberger R. Second order total generalized variation (TGV) for MRI. Magnetic Resonance in Medicine. 2011;65(2):480-91.
9. Wang S, Su Z, Ying L, Peng X, Zhu S, Liang F, Feng D, Liang D, editors. Accelerating magnetic resonance imaging via deep learning. 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI); 2016 2016/04//.
10. Lee D, Yoo J, Tak S, Ye JC. Deep Residual Learning for Accelerated MRI Using Magnitude and Phase Networks. IEEE Transactions on Biomedical Engineering. 2018;65(9):1985-95. doi: 10.1109/TBME.2018.2821699.
11. Liu J, Sun Y, Eldeniz C, Gan W, An H, Kamilov US. RARE: Image Reconstruction Using Deep Priors Learned Without Groundtruth. IEEE Journal of Selected Topics in Signal Processing. 2020;14(6):1088-99. doi:10.1109/JSTSP.2020.2998402.
12. Lehtinen J, Munkberg J, Hasselgren J, Laine S, Karras T, Aittala M, Aila T. Noise2Noise: Learning Image Restoration without Clean Data. arXiv:180304189 [cs, stat]. 2018.
13. M. T. McCann, K. H. Jin, and M. Unser. Convolutional neural networks for inverse problems in imaging. A review, IEEE Signal Process. Mag., vol. 34, no. 6, pp. 85–95, Nov. 2017.
14. A. Lucas,M. Iliadis,R. Molina, and A.K.Katsaggelos Using deep neural networks for inverse problems in imaging: Beyond analytical methods. IEEE Signal Process. Mag., vol. 35, no. 1, pp. 20–36, Jan. 2018.
15. F. Knoll et al., Deep-learning methods for parallel magnetic resonance imaging reconstruction. A survey of the current approaches, trends, and issues. IEEE Signal Process. Mag., vol. 37, no. 1, pp. 128–140, Jan. 2020.
16. D.C. Peters, F.R. Korosec, T.M. Grist, W.F. Block, J.E. Holden, K.K. Vigen, and C.A. Mistretta. Undersampled projection reconstruction applied to MR angiography. Magn. Reson. Med., vol. 43, no. 1, pp. 91–101, 2000.
Figures