2395
Ultra-High Field MRI reveals amyloid plaques-like in a familial Alzheimer’s Disease patient-derived brain organoids1Institut du Cerveau – Paris Brain Institute - ICM, Sorbonne Université, INSERM, CNRS, Paris, France, 2Centre de NeuroImagerie de Recherche – CENIR, Paris, France, 3Molecular Pathophysiology of Parkinson’s Disease Group, Paris, France
Synopsis
This study presents preliminary results obtained on APP muted brain organoids. In vitro images were obtained using ultra-high field MRI and Gd passive staining of samples. MR images were compared to histology and immunohistochemistry slices to confirm the presence of amyloid plaques of mutated APP brain organoid compared to isogenic (control) brain organoid.
Introduction
Cortical organoids are complex stem cell-derived 3D cultures that reproduce features of organization, architecture, and function of the developing brain1. Recent studies have however shown that organoids derived from patients with familial forms of neurodegenerative diseases like Alzheimer’s Disease (AD), Parkinson’s Disease (PD) or Fronto-Temporal Dementia (FTD) could also replicate essential elements of these late-onset conditions, such as abnormal protein aggregation, cellular dysfunction, and neuronal death2–5. Here we present for the first time MR images of brain organoids, that aims to combine the use of ultra-high field with highly-sensitive MR coils to reach high Signal to Noise Ratio (SNR) and Contrast to Noise Ratio (CNR) of small samples with a very high resolution (15 µm isotropic). Our study thus highlight the relevance of brain organoids to study disease mechanisms as well to identify and test novel therapeutic targets.Materials and Methods
Forebrain assembloids were generated from a familial AD patient line (APPV717I “London” mutation) as well as its isogenic control based on previously published protocols6. After 6 months in vitro, the organoids were fixed in 4% PFA then, prior to imaging, were soaked for ~5hours in a mixture PBS and Gd-based contrast agent (Dotarem, Guerbet, France) at a 1:200 ratio. Images were acquired using a Bruker Biopsec 117/16 (Bruker, Germany) equipped with a cryoprobe. Samples were placed on the bottom of the coil in a plastic tube filled with an aprotonic solution (Fluorinert, 3M, USA). Images were acquired with a 3D-GRE sequence with an isotropic resolution of 15 µm with the following parameters: TR/TE = 65/20 ms, Flip Angle = 43°, Nex = 40 , Bandwidth = 50kHz, Mtx = 224*224*176, Scan Time = 28.5h).After imaging, Congo Red stainings as well as anti-Amyloid Beta immunofluorescences were conducted on organoid cryosections (12-30µm thick), and digitized using Zeiss’s Axio Scan Z1.
Results
Dark spots in MR images were identified as beta-amyloid plaques (Figure 1). Image contrast and dark spots were similar to those obtained in APP/PS1 muted mouse using in vivo MRI combined with Gd passive staining techniques7-8. Anti-amyloid beta immunofluorescence (6E10 clone) revealed a strong signal in cortical organoids carrying APPV717I mutations (Figure 2) which was absent from the isogenic controls after 6 months in vitro. This signal, which was also identifiable using Congo Red dye, was strikingly similar to that obtained from the MRI scan.Discussion and Conclusions
To our knowledge, this is the first time that MR Images of brain organoids are reported, especially using a familial AD patient line. This proof of concept experiment may increase the translational value of the organoid model for studying the cellular responde to immunotherapies.Future experiments will require to use time-matched isogenic controls. We further need to implement registration methods of MRI scans to 3D immunofluorescences of the same organoids to fully confirm the signal overlap of amyloid plaques. Furthermore, this preliminary study might serve as a fisrt step to bridge the gap between imaging, neuropathology and neuropharmacology of complex 3D stem-cell derived culture.
Acknowledgements
This work was supported by MSD avenir ‘miniAD MSD Avenir’References
1. Lancaster, M. A. & Knoblich, J. A. Organogenesisin a dish: Modeling development and disease using organoid technologies. Science 345, (2014).
2. Gonzalez, C. et al. Modeling amyloid beta and tau pathology in human cerebral organoids. Molecular Psychiatry 2363–2374 (2018) doi:10.1038/s41380-018-0229-8.
3. Zhao, J. et al. APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat Commun 11, 5540 (2020).
4. Bowles, K. R. et al. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell 184, 4547-4563.e17 (2021).
5. Ahfeldt, T. et al. Pathogenic Pathways in Early-Onset Autosomal Recessive Parkinson’s Disease Discovered Using Isogenic Human Dopaminergic Neurons. Stem Cell Reports 14, 75–90 (2020).
6. Sloan, S. A., et al. Generation and assembly of human brain region–specific three-dimensional cultures. Nature Protocols 13, 2062–2085 (2018).
7. Santin M.D. et al. In vivo detection of amyloid plaques by gadolinium-stained MRI can be used to demonstrate the efficacy of an anti-amyloid immunotherapy”, Frontier in aging Neuroscience, 2016.
8. Santin M. D. et al. Fast in
vivo Imaging of amyloid plaques using µ-MRI Gd-staining combined with
ultrasound-induced blood brain barrier opening", Neuroimage (79), Oct.
2013.
Figures
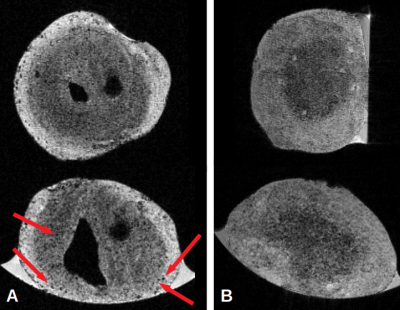
A: Coronal and Axial MR Images of an APPV717I brain organoid. Red arrows indicate the presence of dark spots identified as amyloid plaques. Voxel size = 15*15*15 µm3
B: Coronal and Axial MR Images of an isogenic brain organoid. no dark spots were clearly identified as amyloid plaques on these images. Voxel size = 15*15*15 µm3
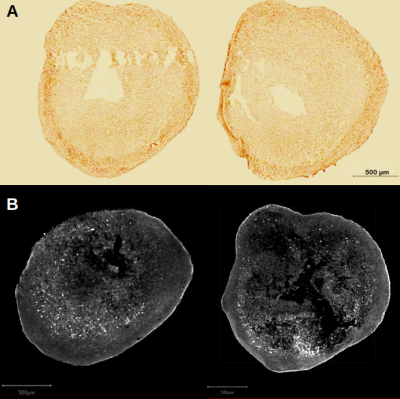
A: Histological slices of the APPV717I brain organoid (12 µm sections). Red coloration confirms the presence of beat-amyloid deposits.
B: Immunofluorescence slices of anti beta-amyloid with a 6E10 clone (30 µm sections). White dots indicates the presence of amyloid plaques.