2333
Imaging ex-vivo cynomolgus monkey brain in 150 um isotropic resolution1Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of, 2National Primate Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Cheongju, Korea, Republic of, 3Department of Functional Genomics, KRIBB School of Bioscience, Korea University of Science and Technology (UST), Daejeon, Korea, Republic of, 4Center for Research Equipment, Korea Basic Science Institute, Cheongju, Korea, Republic of, 5Futuristic Animal Resource & Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Cheongju, Korea, Republic of
Synopsis
Due to the small size of the cynomolgus brain, imaging the fine structures in the brain is a challenging task. In this study, a sample preparation method is suggested for imaging the ex-vivo cynomolgus monkey brain with sub-millimeter resolution in 7T MRI. Using the proposed method, we acquired 150 um isotropic resolution MR images for whole brain, and the fine structures in the cynomolgus monkey brain was observed.
Introduction
The cynomolgus monkey (macaca fascicularis) is one of the most commonly used experimental animals, including its usage as an animal model for Parkinson’s disease.1 However, due to its small-sized brain,2 observing the detailed structures can be a challenging task. Using a 7T MRI is a good solution to reach high resolution leveraging increased MR signal power, but, arising problems regarding system imperfection (e.g. imperfect B0 shimming) are more severe than 3T, degrading the image quality. In this study, we proposed a sample preparation method for acquiring sub-millimeter resolution MR images of the ex-vivo cynomolgus monkey brain. With the proposed method, the whole-brain T2*-weighted image and QSM image were acquired with 150 um isotropic resolution, and the structural details were explored and compared to the histology images from references.3Methods
[Ex-vivo sample preparation]An ex-vivo brain of the cynomolgus monkey with a history of Parkinson’s disease was prepared for this study. For the preparation of the brain sample, three steps were performed. They were 1) designing the unique shape of the container for better B0 shimming, 2) reducing bubbles for artifact reduction, and 3) agarose embedding for reducing motions.
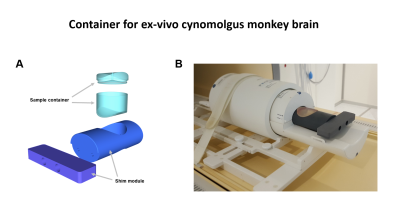
Firstly, a container was designed to improve B0 shimming performance. Figure 1 shows the 3D image and demonstration images of the designed container. The container consists of the sample container and the shim module. For experiments, the brain sample is placed in the sample container, and the sample container is placed in the shim module. The container was designed to fit the size of the 16-channel human wrist coil, which is used for the following experiments.
Next, bubbles were removed in the sample container. Because the material around the bubble and the air inside the bubble have different susceptibility values, it may induce susceptibility artifacts. We tried to remove the small bubbles existing in samples using a vacuum chamber.
Thirdly, the brain sample was embedded using an agarose gel. Because vibrations can be generated by the machine during an MRI scan, the brain sample may be prone to unexpected motions. Therefore, to fix the position of the brain sample in the sample container, agarose gel was used for embedding the brain sample.
[Data acquisition & image processing]
With the proposed sample preparation method, the ex-vivo sample was scanned at 7T Philips MRI with the 16-channel human wrist coil. The 3D multi-echo gradient-recalled echo (GRE) scan was performed with the following scan parameters: voxel size = 150 um isotropic, TE1/ΔTE/TR = 5.5/4.7/100 ms, # of echoes = 4, flip angle = 24°, and total scan time = 4.2 hours. From the complex images of DICOM data, the magnitude image was generated by the square root of the sum of squares over echoes, and the QSM map was generated by a series of post-processing steps including Laplacian phase unwrapping, HARPERELLA background field removal, and iLSQR. The magnitude image and QSM image were compared with histology image to explore that the MR images can provide the fine structure in the brain sample.
Results
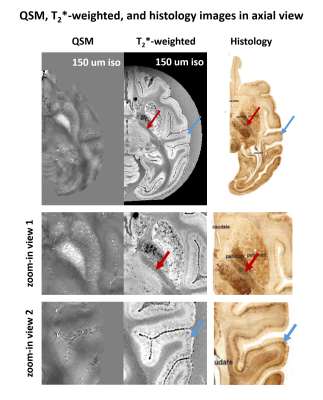
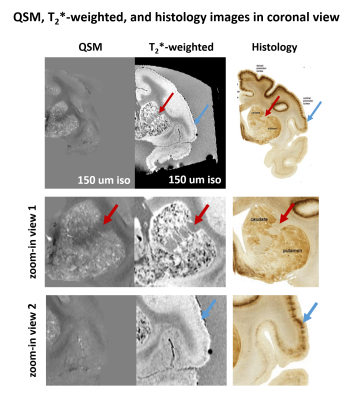
Figure 2 shows the QSM, T2*-weighted, and histology images in the axial view of a representative slice. Comparing the T2*-weighted image and the histology image, the unique structure of the internal capsule and laminar structure of the cortex was consistently observed in both images. However, they were not observed in QSM.Figure 3 shows the QSM, T2*-weighted, and histology images in the coronal view of a representative slice. The unique structure between caudate and putamen was consistently observed in QSM, T2*-weighted, and histology images. Also, the laminar structure of the cortex was consistently observed both in T2*-weighted and histology images.
Discussion and Conclusion
In summary, we proposed a sample preparation method for acquiring sub-millimeter resolution images of the ex-vivo cynomolgus monkey brain. By acquiring high resolution images, the detailed structure in the brain can be observed even for the small-sized brain. When comparing MR images with the reference histology images, MR images contain abundant information on histological structures.Acknowledgements
This work has been supported by Heuron Co. Ltd. and by the Korea government (MSIT) (No.NRF-2021R1A2B5B03002783)References
1. Dauer, W. & Przedborski, S. Parkinson’s Disease: Mechanisms and models. Neuron 39, 889–909 (2003).
2. Herculano-Houzel, S. The human brain in numbers: A linearly scaled-up primate brain. Front. Hum. Neurosci. 3, 1–11 (2009).
3. Saleem, K. S. K. & Logothetis, N. K. N. A combined MRI and histology Atlas of the rhesus monkey brain in stereotaxic coordinates. Academic Press, London (2007).
Figures