2286
Isotropic resolution volumetric liver T2 mapping using a free-breathing navigator-gated radial stack-of-stars T2-prepared Dixon acquisition1Department of Diagnostic and Interventional Radiology, School of Medicine, Technical University of Munich, Munich, Germany, 2Philips GmbH Market DACH, Hamburg, Germany
Synopsis
Volumetric liver T2-mapping is of interest in the characterization of focal lesions and diffuse disease but remains technically challenging due to respiratory motion. The present work introduces a novel T2-prepared (T2prep) radial stack-of-stars two-point Dixon gradient echo sequence (T2prep-SoS-Dixon-TFE) for achieving motion-robust 3D isotropic T2-mapping in navigator-gated scans using a B0/B1-insusceptible modified adiabatic BIR-4 RF-pulse for T2prep. T2-mapping is performed via dictionary matching to Bloch simulations for the Dixon-decomposed water signal. The proposed method enables motion-robust liver fat confounder-free volumetric T2 quantification in better agreement with MRS compared to GraSE and may improve the clinical applicability of free-breathing abdominal T2-mapping.
Purpose:
Volumetric T2-weighted imaging is of clinical interest in the characterization of focal liver lesions[1] and T2 mapping has shown promising results in the staging of liver fibrosis[2,3]. However, 3D T2-weighted imaging and T2 mapping of the liver remains challenged by the strong sensitivity of the region to respiratory motion and therefore imaging has been frequently performed falling back to 2D acquisitions and breath-hold scans[4]. Stack-of-stars (SoS) acquisitions allow volumetric coverage and have gained popularity for liver imaging due to its robustness to motion and self-navigation capabilities[5-8]. Recent works presented frameworks for inversion-recovery-prepared radial SoS imaging and demonstrated its feasibility for volumetric T1 and T2 mapping in the liver during free breathing[9-13] but did not assess the agreement of T2 mapping to reference measurements. A T2-prepared (T2prep) radial stack-of-stars two-point Dixon gradient echo sequence (T2prep-SoS-Dixon-TFE) technique was also proposed for the acquisition of isotropic resolution volumetric liver T2-weighted imaging and T2 mapping based on navigator-gated scans[14]. In this work, we refine the T2prep-SoS-Dixon-TFE methodology, investigate its advantages over gradient-spin-echo (GraSE) -based T2 mapping in volunteers and phantoms in comparison to single-voxel MRS reference measurements.Methods:
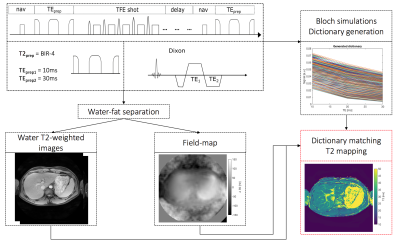
Pulse Sequence:An adiabatic T2prep module based on a modified BIR-4 RF-pulse[14-17] was combined with a dual-echo radial SoS TFE readout (Fig.1). T2 weighting was modulated by changing the gap time between the BIR-4 segments. The two-point Dixon data was used for water-fat separation and field-map estimation (Fig.1). T2 maps were obtained by dictionary matching of the water signal with and without taking the field-map into consideration for higher accuracy of the T2 mapping (Fig.1).
Dictionary generation:
The signal evolutions dictionary was created using Bloch simulations for TEprep=10,30ms, a fixed T1 of 800ms, T2 from 10ms to 100ms with a step size of 2ms and a field-map range from -300Hz to 300Hz with a step size of 20Hz.
In vivo - liver and phantom relaxometry:
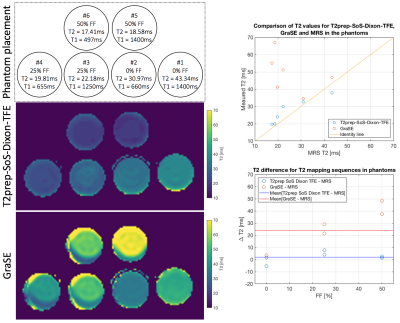
The proposed navigator-gated T2prep-SoS-Dixon-TFE sequence was acquired in six phantoms with varying fat fraction (FF), T1, T2 (Fig.2) and in twelve healthy volunteers. In a subset of four volunteers, the sequence was acquired twice (with subject repositioning) to estimate reproducibility. Sequence parameters were as follows: FOV: 400x400x200mm3, acquisition voxel size: 3mm3 isotropic, TR/TE1/TE2 = 3.8/1.11/2.2ms, TFE angle=8° and shot duration: 550ms, effective duration of the T2prep: 10/30ms, nominal scan time: 3m26s. For water-fat-decomposition, the vendor’s implementation was used[18]. For comparison, the vendor’s multi-echo GraSE T2 mapping sequence was acquired (FOV: 400x400x200mm3, acquisition voxel size: 3mm isotropic, 8 echoes, TR=1641ms, TEs=16ms to 64ms in 8ms steps, scan time 2m24s) using respiratory triggering and a single-voxel multi-TE STEAM MRS (TR/TM=1500/16ms, VOI=15x15x15mm3, 1 average, TEs=10ms to 50ms in 10ms steps) acquired in an 8s breath-hold were performed in the liver tissue. A single-voxel multi-TE STEAM MRS with the same parameters was acquired in each phantom.T2 values from the imaging sequences were calculated based on a voxel-wise dictionary matching of the water T2-weighted images including and not including the measured field-map from the Dixon approach. MRS data were processed with in-house MATLAB scripts including zero-order phasing, Gaussian apodization, frequency alignment and weighted coil-combination. T2 values for water and fat were obtained by joint peak area quantification for all TEs in the frequency domain.
Results
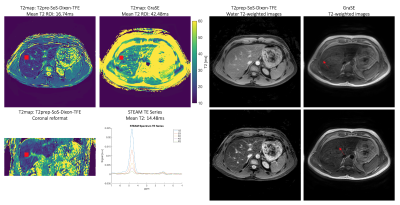
Figure 2 shows the T2 mapping results of the phantom study for T2prep-SoS-Dixon-TFE and GraSE. GraSE-based T2 maps showed chemical shift displacement artifacts and large T2 variations in the presence of fat.In Figure 3 obtained in vivo T2 maps for GraSE and T2prep-SoS-Dixon-TFE are displayed together with matching spectra and T2-weighted maps. Using the MRS as a reference measurement, the GraSE led to an overestimation of 28ms whereas the T2prep-SoS-Dixon-TFE only overestimated T2 by 2.3ms. The GraSE T2 maps and T2-weighted images showed overall higher sensitivity to motion.
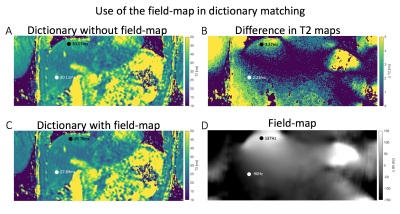
Figure 4 shows dictionary matching-based T2 mapping results with and without using information from the Dixon-based field-map. Use of the field-map reduced T2 overestimation of the T2prep-SoS-Dixon-TFE, especially in regions with strong field-map variations close to the lungs.
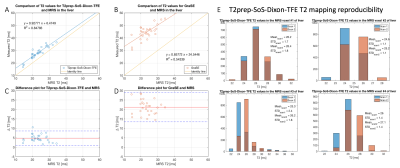
Figure 5 shows the comparison of T2 values between T2prep-SoS-Dixon-TFE, GraSE and MRS for liver scanning. Both T2prep-SoS-Dixon-TFE and GraSE tend to overestimate T2. However, the T2prep-SoS-Dixon-TFE showed a better agreement with MRS (slope=0.93, intercept=6.4ms, R2=0.85) than GraSE with MRS (slope=0.86, intercept=24.5ms, R2=0.54). T2prep-SoS-Dixon-TFE showed good reproducibility with the difference in T2 quantification of not more than 1.1ms between scans (Fig.5E).
Discussion & Conclusion
The present work introduced a novel T2prep-SoS-Dixon-TFE method for motion robust 3D isotropic resolution T2 mapping and demonstrated its feasibility in phantoms and in vivo in the liver with measured T2 values in good agreement with MRS. Incorporating the field-map information in the dictionary matching further improved T2 accuracy. In conclusion, the proposed method constitutes a promising tool for motion-robust volumetric isotropic resolution T2-weighted imaging and T2 mapping in the liver.Acknowledgements
The present work was supported by the TUM International Graduate School of Science and Engineering (TUM-ICL Joint Academy of Doctoral Studies). The authors also acknowledge research support by Philips Healthcare.References
[1] Keerthivasan MB, Galons JP, Johnson K, Umapathy L, Martin DR, Bilgin A, Altbach MI. Abdominal T2‐Weighted Imaging and T2 Mapping Using a Variable Flip Angle Radial Turbo Spin‐Echo Technique. Journal of Magnetic Resonance Imaging 2021.
[2] Hoffman D.H., Ayoola A., Nickel D. et al. T1 mapping, T2 mapping and MR elastography of the liver for detection and staging of liver fibrosis. Abdom Radiol 45, 692–700 (2020). https://doi.org/10.1007/s00261-019-02382-9
[3] Guimaraes AR, Siqueira L, Uppal R, Alford J, Fuchs BC, Yamada S, Tanabe K, Chung RT, Lauwers G, Chew ML, Boland GW, Sahani DV, Vangel M, Hahn PF, Caravan P. T2 relaxation time is related to liver fibrosis severity. Quant Imaging Med Surg. 2016 Apr;6(2):103-14. doi: 10.21037/qims.2016.03.02. PMID: 27190762; PMCID: PMC4858462.
[4] Fahlenkamp UL, Ziegeler K, Adams LC, Böker SM, Engel G, Makowski MR. Native T1 mapping for assessment of the perilesional zone in metastases and benign lesions of the liver. Sci Rep. 2020 Jul 30;10(1):12889. doi: 10.1038/s41598-020-69819-w. PMID: 32733016; PMCID: PMC7393097.
[5] Chandarana H, Block TK, Rosenkrantz AB, Lim RP, Kim D, Mossa DJ, Babb JS, Kiefer B, Lee VS. Free-breathing radial 3D fat-suppressed T1-weightedgradient echo sequence: a viable alternative for contrast-enhanced liver imaging in patients unable to suspend respiration. Investigative radiology2011;46(10):648-653.
[6] Benkert T, Mugler JP, 3rd, Rigie DS, Sodickson DK, Chandarana H, Block KT. Hybrid T2 - and T1 -weighted radial acquisition for free-breathingabdominal examination. Magn Reson Med 2018;80(5):1935-1948.
[7] Benkert T, Feng L, Sodickson DK, Chandarana H, Block KT. Free-breathing volumetric fat/water separation by combining radial sampling, compressed sensing, and parallel imaging. Magn Reson Med 2017;78(2):565-576.
[8] Feng L, Grimm R, Block KT, Chandarana H, Kim S, Xu J, Axel L, Sodickson DK, Otazo R. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med 2014;72(3):707-717.
[9] Feng L, Block KT, Benkert T, Tian Y, Liu C, Liu F, Fayad Z, Yang Y. Stack-of-Stars Inversion-Recovery MRI for Free-Breathing T1 Mapping and IR-PreparedFat/Water Separation, Proc ISMRM 2020, p 1015
[10] Feng L, Liu F, Soultanidis G, Liu C, Benkert T, Block KT, Fayad Z, Yang Y. Magnetization-prepared GRASP MRI for rapid 3D T1 mapping and fat/water-seperated T1 mapping. Magn Reson Med 2021;81(1):97-114
[11] Milotta G, Cruz G, Neji R, Prieto C, Botnar R. 3D Isotropic-resolution Non-rigid Morion Compensated Liver T1, T2 and fat fraction mapping. Proc. Intl. Soc. Magn. Reson. Med. 29 (2021), 1234
[12] Wang N, Cao T, Han F, Xie Y, Zhong X, Ma S, Zhang X, Bi X, Noureddin M, Deshpande V, Christodoulou AG, Li D. Six-Dimensional, Free-Breathing Multitasking Multi-Echo (MT-ME) MRI for Whole-Liver T1, PDFF, and R2* Quantification. Proc. Intl. Soc. Magn. Reson. Med. 29 (2021), 0609
[13] Muslu Y, Cashen TA, Mandava S, Reeder SB. Free-Breathing, Confounder Corrected T1 Mapping in the Liver with Stack-of-Stars Inversion Recovery MRI.Proc. Intl. Soc. Magn. Reson. Med. 29 (2021), 0611
[14] Zamskiy M, Weidlich D, Weiss K, Makowski M, Braren R, Karampinos DC. Isotropic resolution volumetric liver T2 weighted imaging and T2 mapping using a navigator-dated radial stack-of-stars T2 prepared acquisition. Proc. Intl. Soc. Magn. Reson. Med. 29 (2021), 1875
[15] Weidlich D, Schlaeger S, Kooijman H, Börnert P, Kirschke JS, Rummeny EJ, Haase A, Karampinos DC. T2 mapping with magnetization-prepared 3D TSE based on a modified BIR-4 T2 preparation. NMR Biomed. 2017 Nov;30(11). doi: 10.1002/nbm.3773. Epub 2017 Aug 4. PMID: 28777496.
[16] Klupp E, Weidlich D, Schlaeger S, Baum T, Cervantes B, Deschauer M, Kooijman H, Rummeny EJ, Zimmer C, Kirschke JS, Karampinos DC. B1-insensitive T2 mapping of healthy thigh muscles using a T2-prepared 3D TSE sequence. PLoS One. 2017 Feb 14;12(2):e0171337. doi: 10.1371/journal.pone.0171337. PMID: 28196133; PMCID: PMC5308846.
[17] Ruschke S, Weidlich D, Diefenbach M, Eggers H, Kooijman H, Hu HH, Rummeny EJ, Haase A, Kirschke JS, Baum T, Karampinos DC. Simultaneous T2, T2’and PDFF mapping in the spine using an adiabatic T2-prepared time-interleaved multi-echo gradient echo acquisition. Proc. Intl. Soc. Magn. Reson. Med. 24 (2016), 0568
[18] Eggers H, Brendel B, Duijndam A, Herigault G. Dual-echo Dixon imaging with flexible choice of echo times. Magn Reson Med. 2011 Jan;65(1):96-107. doi: 10.1002/mrm.22578.
Figures