2248
Quantifying Relative Displacement and Phase at the Skull-Brain Interface using MR Elastography1Biomedical Engineering, University of Delaware, Newark, DE, United States, 2Henry M. Jackson Foundation, Bethesda, MD, United States, 3National Institutes of Health, Bethesda, MD, United States
Synopsis
This study uses a novel multishot spiral magnetic resonance elastography (MRE) pulse sequence to simultaneously measure skull and brain motion in vivo in a single acquisition. Results in a healthy volunteer and a fat-water phantom showed a reduced transmission of motion from the skull to the brain in all three directions of motion. Additionally, the skull and brain appear to be out of phase in motion in both the x (left-right) and z (superior-inferior) directions. This research provides a basis for future studies quantifying skull-brain coupling including with multiple actuation frequencies and excitation directions.
Introduction
Traumatic brain injury (TBI) is a leading cause of death and disability1,2 that results from a direct impact or blast when the force applied to the head exceeds the protection provided by the skull-brain interface3. Despite the prevalence and severity of TBI, the brain’s response to trauma and the mechanisms involved with injury remain unclear. Quantifying the relative motion between the brain and skull at this interface can help researchers better understand how skull acceleration leads to brain injury. Data on skull-brain coupling can be used to improve computational TBI models that will elucidate these mechanisms and aid in the design of protective equipment4. Magnetic resonance elastography (MRE) can measure brain deformation noninvasively to estimate brain mechanical properties but can also be used to measure the relative motion of the skull and brain in vivo5,6. Previous work by Yin et al. introduced a method to separately image skull and brain by isolating signal from fat in the bone marrow3. In this work, we expand on that approach using a modified multishot spiral MRE sequence to simultaneously measure both skull and brain motion to ultimately quantify the transmission of motion from the skull to the brain.Methods
Imaging: MRE displacement data was acquired using a modified 2D multishot spiral MRE sequence7. We used a 12-shot constant-density spiral sequence with no fat suppression to see the fat signal while minimizing blurring through short data readout times. Imaging parameters included: 240 x 240 mm field-of-view, 2 mm isotropic resolution, 20 slices, TR/TE = 2000/56 ms, and 4 phase offsets. All scanning was conducted using a Siemens 3T Prisma MRI scanner and a 64-channel head coil. Vibrations were generated at 50 Hz and 100 Hz using a Resoundant pneumatic actuation system (Rochester, MN) and a passive pillow driver. Low motion encoding gradient strength was used to capture rigid body motion while avoiding temporal phase wrapping3,5.Phantom: A layered fat-water phantom was constructed for sequence development to mimic signal from the brain and skull. Two nested bowls were used to separate the fat and water components and to represent the general shape of the skull and brain. The fat layer was intended to mimic bone marrow in the brain with a fat fraction of 50% and 3 wt.% agar which causes this layer to be very stiff. Peanut oil was used as the fat component due to its similar NMR spectrum to in vivo fat8. The water-based portion representing the brain was created with a 0.8 wt.% agar solution9. The phantom was imaged at both 50 Hz and 100 Hz vibration frequencies.
In vivo: A single human subject (female, age 23) was scanned using the fat-water MRE sequence at both 50 Hz and 100 Hz vibration frequencies.
Analysis: Imaging data was iteratively reconstructed10. Phase images were processed to obtain maps of complex motion with sensitization in the x (left-right), y (anterior-posterior), and z (superior-inferior) directions. Representative masks were created at the interface of the skull and brain (Figure 1). Points around the circumference of the skull were defined in terms of polar coordinates and described by their polar angle, θ. Displacement amplitude and relative phase of the harmonic motion was quantified for each point in both skull and brain.
Results
Our in vivo analysis of brain and skull motion showed an overall lower displacement amplitude of the brain relative to the skull, indicating reduced transmission of motion to the brain in all three directions (Figure 2). The skull motion amplitude appears to follow a sinusoidal pattern for each individual direction starting from the site of actuation and represents the rigid body motion in three dimensions. The brain shows a similar pattern but at a lower amplitude than the skull. The phase plots show that the skull and brain are moving out of phase in both the x (left-right) and z (superior-inferior) directions while the brain and skull in the y direction (anterior-posterior) appear to remain in phase. At the higher frequency of 100 Hz, the brain appears to move less than at 50 Hz (Figure 3). We see similar trends in the phantom, however the fat layer and agarose layer appear to move with more similar amplitudes of motion, and in some cases with the agarose displacing more than the fat layer, possibly caused by sloshing from the agar material having a free surface (Figures 4-5).Discussion & Conclusions
These preliminary in vivo results support the hypothesis that the brain moves out of phase with the skull as captured by our modified fat-water spiral MRE sequence. Further sequence developments will seek to optimize the skull signal and reduce contamination from the scalp6. Future studies will include developing a new phantom to better mimic the interface between the brain and skull and validating the relative motion measurements. An increased range of actuation frequencies with multiple excitation directions will allow us to comprehensively quantify skull-brain transmission. Understanding this relationship will allow researchers to improve TBI models by accounting for the skull-brain interface and evaluate how this relative motion changes in patients with a history of mild TBI.Acknowledgements
This project was supported by NIH grant U01-NS112120 and the NIH Bench-to-Bedside program.References
1. Hyder, A. A., Wunderlich, C. A., Puvanachandra, P., Gururaj, G. & Kobusingye, O. C. The impact of traumatic brain injuries: A global perspective. NeuroRehabilitation 22, 341–353 (2007).
2. Nguyen, R. et al. The International Incidence of Traumatic Brain Injury: A Systematic Review and Meta-Analysis. Can. J. Neurol. Sci. 43, 774–785 (2016).
3. Yin, Z. et al. In vivo characterization of 3D skull and brain motion during dynamic head vibration using magnetic resonance elastography. Magn. Reson. Med. 80, 2573–2585 (2018).
4. Bayly, P. V. et al. MR Imaging of Human Brain Mechanics In Vivo: New Measurements to Facilitate the Development of Computational Models of Brain Injury. Ann. Biomed. Eng. 2021 1–16 (2021) doi:10.1007/S10439-021-02820-0.
5. Badachhape, A. A. et al. The Relationship of Three-Dimensional Human Skull Motion to Brain Tissue Deformation in Magnetic Resonance Elastography Studies. J. Biomech. Eng. 139, 0510021 (2017).
6. Badachhape, A. A., Okamoto, R. J., Johnson, C. L. & Bayly, P. V. Relationships between scalp, brain, and skull motion estimated using magnetic resonance elastography. J. Biomech. 73, 40–49 (2018).
7. Johnson, C. L. et al. Magnetic resonance elastography of the brain using multishot spiral readouts with self-navigated motion correction. Magn. Reson. Med. 70, 404–412 (2013).
8. Bush, E. C. et al. Fat-Water Phantoms for Magnetic Resonance Imaging Validation: A Flexible and Scalable Protocol. J. Vis. Exp 57704 (2018) doi:10.3791/57704.
9. McIlvain, G. et al. Reliable preparation of agarose phantoms for use in quantitative magnetic resonance elastography. J. Mech. Behav. Biomed. Mater. 97, 65–73 (2019).
10. Sutton, B. P., Noll, D. C. & Fessler, J. A. Fast, iterative image reconstruction for MRI in the presence of field inhomogeneities. IEEE Trans. Med. Imaging 22, 178–188 (2003).
Figures
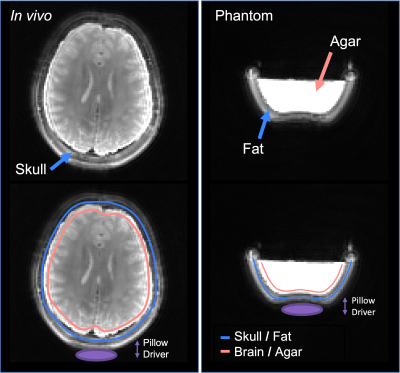
Figure 1. Magnitude images obtained from adapted multishot spiral MRE sequence in a phantom and in vivo. Both water and fat components are visible, and masks of brain and skull components used for quantification are overlaid.
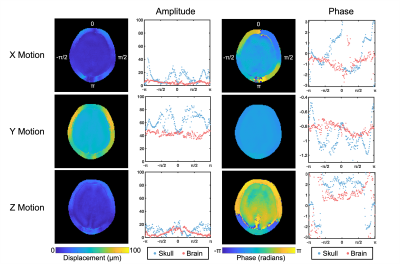
Figure 2. Amplitude and phase of motion in the x, y, and z directions at 50 Hz in vivo showing relative differences between skull and brain motion.
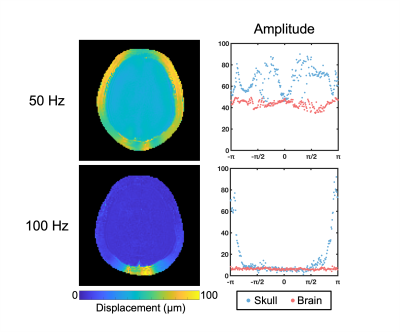
Figure 3. Total amplitude displacement of the skull and brain at 50 Hz and 100 Hz in vivo.
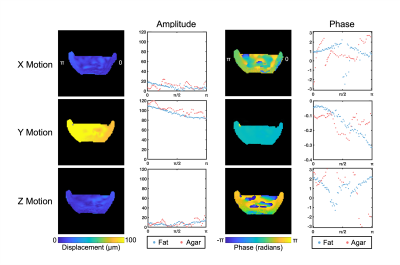
Figure 4. Amplitude and phase of motion in the x, y, and z directions at 50 Hz in the fat-water phantom showing relative differences between fat and agar motion.
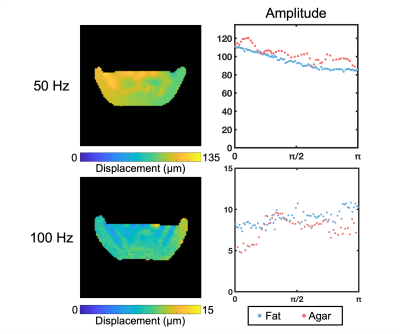
Figure 5. Total amplitude displacement of the fat and agar components at 50 Hz and 100 Hz in the fat-water phantom.