2235
Cascaded parallel imaging for 3D deep learning reconstruction1UIH America, Inc., Houston, TX, United States, 2United Imaging Intelligence, Cambridge, MA, United States
Synopsis
This study demonstrates the effects on image reconstruction by integrating a separate parallel imaging layer with the deep learning module. With such additional layer, deep learning reconstruction is flexible and reliable for highly under-sampled data.
Introduction
Multi-parametric MRI techniques have been explored recently. [1-5] The challenges of multi-parametric imaging still lie in achieving fast 3D high-resolution imaging capacity. Since the compressed sensing (CS) algorithms are often iterative and time-consuming, it is difficult for CS-based method to reconstruct such a large amount of high-resolution 3D data within a clinically acceptable duration. Recently, ReconNet3D network [6] has been developed to reconstruct multi-channel 3D images by processing individual channel separately. Reconstructed images from all channels were sent to MDI module [5, 8] for multi-parametric imaging. However, when acceleration factor is high, the acquired image quality is degraded. 3D Parallel Imaging with transitional Auto-calibration (3D-PITA) [7] have been used for highly accelerated 3D imaging. In this work, a k-space based parallel imaging layer is cascaded with state-of-the-art ReconNet3D network. We perform an initial study on the cascaded reconstruction strategy of highly under-sampled (R=4) data.Methods
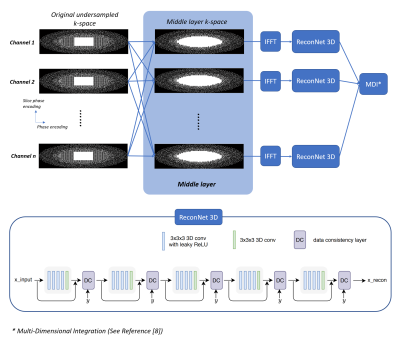
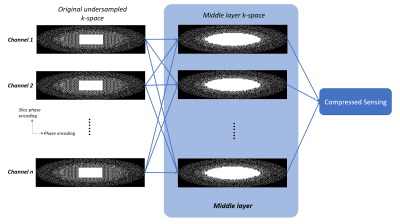
k-Space data is under-sampled along ky and kz directions. The autocalibration signal region is fully sampled. The high frequency region is under-sampled using Poisson-Disc pattern with an oval mask. A transition region is designed in between, with 2.67-fold acceleration (a separate sampling pattern as shown in Fig. 1). Using such a sampling pattern, the calibration and reconstruction can be performed separately in a middle layer as shown in Fig. 1. The middle layer works as a Multiple channel In Multiple channel Out (MIMO) system. With such middle layer, unacquired k-space data in transition region is calculated. A cascaded CNN model with 3D convolutional layers and data consistency layers (named ReconNet3D) [6] is connected to each channel of the middle layer. ReconNet3D was designed to reconstruct the multi-channel image by processing individual channel separately (i.e. without any a priori knowledge of signals from any other channels), and it was trained and tested as such. One concern is that whether the middle layer introduces additional noise. To address this concern, a second reconstruction pipeline is constructed as shown in Fig. 2. A compressed sensing reconstruction unit is connected to the middle layer. The middle layer can be switched on or off. The two pipelines were implemented on the inline image reconstruction pipeline of a 3T scanner (uMR890, United Imaging Healthcare, Shanghai, China).Results and discussion
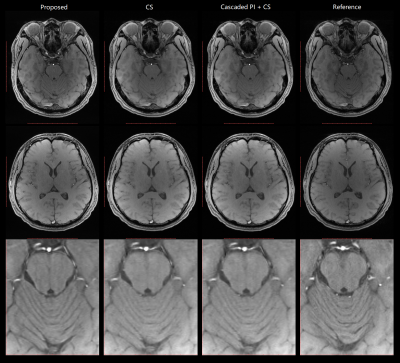
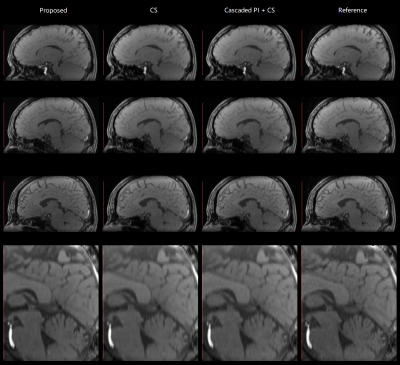
The cascaded parallel imaging with ReconNet3D model was tested via volunteer brain scans, using the 32-channel head coil of the scanner and a 3D GRE sequence. TR/TE=20/3.75ms, FA=15°, voxel size = 1x1x3mm3. Total scan time was 1min 40s. Unacquired k-space data of each channel were first reconstructed by the middle layer, and then individually by the ReconNet3D model and subsequently combined using sum-of-square (SOS). The Cascaded parallel imaging with CS was tested, using retro reconstruction of the same dataset, through Pipeline 2. CS reconstruction without the middle layer is also tested, using retro reconstruction of the same dataset, through Pipeline 2 with middle layer OFF.A new scan for reference was performed with the same slice position and same key imaging parameters, but with 1.68x acceleration inline parallel imaging protocol. The scan time was 3 minutes 57s. Fig. 3 and Fig 4 compare images from the proposed (accelerated Cascaded PI + ReconNet3D) method, CS reconstruction, CS reconstruction with middle layer, and the reference images. The image quality from either Cascaded PI + ReconNet3D, CS, cascaded PI + CS were generally good, and were consistent with reference images. The Cascaded PI + ReconNet3D maintained sharper details, compared with CS reconstructions. The image quality from CS with middle layer on and off are similar, which suggests additional noise from middle layer is negligible. The image peak signal-to-noise ratio (PSNR) of Cascaded PI + ReconNet3D, CS, and cascaded PI + CS were 16.43, 17.81, 17.82, respectively, which also indicate that noise level of middle layer is negligible.
Conclusion
In conclusion, we have demonstrated the reliability of cascaded parallel imaging layer for deep learning model (ReconNet3D) on highly accelerated image reconstruction using the same dataset. Images from the ReconNet3D have richer details than compressed sensing.Acknowledgements
No acknowledgement found.References
[1] Chen, Yongsheng, et al. "STrategically Acquired Gradient Echo (STAGE) imaging, part I: Creating enhanced T1 contrast and standardized susceptibility weighted imaging and quantitative susceptibility mapping." Magnetic resonance imaging 46 (2018): 130-139.
[2] Wang, Yu, et al. "STrategically Acquired Gradient Echo (STAGE) imaging, part II: Correcting for RF inhomogeneities in estimating T1 and proton density." Magnetic resonance imaging 46 (2018): 140-150.
[3] Haacke, E. Mark, et al. "STrategically Acquired Gradient Echo (STAGE) imaging, part III: Technical advances and clinical applications of a rapid multi-contrast multi-parametric brain imaging method." Magnetic resonance imaging 65 (2020): 15-26.
[4] Tanenbaum, Lawrence N., et al. "Synthetic MRI for clinical neuroimaging: results of the magnetic resonance image compilation (MAGiC) prospective, multicenter, multireader trial." American Journal of Neuroradiology 38.6 (2017): 1103-1110.
[5] Ye, Yongquan, et al. "MULTI‐parametric MR imaging with fLEXible design (MULTIPLEX)." Magnetic Resonance in Medicine (2021).
[6] Chen, Eric Z., et al. "Accelerating 3D MULTIPLEX MRI Reconstruction with Deep Learning." arXiv preprint arXiv:2105.08163 (2021).
[7] He, Ren. 3D Parallel Imaging with Transition Auto-Calibration (3D-PITA). Diss. State University of New York at Buffalo, 2015.
[8] Ye, Yongquan, et al. "A multi‐dimensional integration (MDI) strategy for MR T2* mapping." NMR in Biomedicine (2021): e4529.
Figures