2233
Deep learning-based segmentation of the parasagittal dural space from non-contrast anatomical MRI image: a resource for glymphatic studies1Neurology, Vanderbilt University Medical Center, Nashville, TN, United States, 2Radiology and Radiological Sciences, Vanderbilt University Medical Center, Nashville, TN, United States, 3Psychiatry and Behavioral Sciences, Vanderbilt University Medical Center, Nashville, TN, United States
Synopsis
The overarching goal of this work is to develop and validate novel deep learning algorithms for segmenting the parasagittal dural (PSD) space, which has been hypothesized to harbor cerebral lymphatic channels, from standard non-contrast anatomical imaging. Specifically, contrasted based MRI studies have recently suggested that the PSD may be important for CSF clearance, however existing methods for evaluating this space require administration of exogenous contrast and time-consuming manual tracing, thereby limiting generalizability. We propose a new segmentation method using non-contrasted MRI, and we validate this method in a mixed cohort of older adults with and without neurodegeneration.
Introduction
The overarching goal of this work is to develop and validate novel deep learning algorithms for segmenting the parasagittal dural (PSD) space, which has been hypothesized to harbor cerebral lymphatic channels, from standard non-contrast anatomical imaging. More specifically, recent studies have emphasized the importance of a human glymphatic system for metabolic waste and protein clearance1, whereby cerebrospinal fluid (CSF) moves from the subarachnoid space to a defined region surrounding the dural sinuses. While it remains incompletely characterized, T1-weighted imaging after exogenous intrathecal contrast injection has shown that the PSD space demonstrates contrast dynamics consistent with trans arachnoid fluid passage. These findings support a potential fundamental role of this region for glymphatic clearance. However, limited approaches are available for quantifying this space non-invasively in vivo. Recent works proposed to study this space using contrast imaging combined with manual segmentation, or standard threshold methods2,3. While this work has improved our understanding of this region, developments are needed to improve feasibility through automation of non-contrasted scans, while also improving accuracy and reproducibility. Here, we develop a novel segmentation method to extract the PSD using only one non-invasive anatomical MRI sequence. We hypothesize that the PSD volume can be accurately segmented using this algorithm, which we evaluate by comparing outputs with gold-standard manual tracing from a board-certified neuroradiologist in a mixed cohort of individuals with and without neurodegeneration.Methods
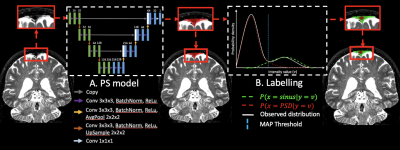
Participants (Table 1; n=19) provided informed, written consent and were scanned at 3T (Philips Healthcare) with dual body coil radiofrequency transmission and 32-channel SENSE phased array reception. Acquisition. 3D T2-weighted sagittal VISTA, echo time=310ms, repetition time=2.7ms, flip angle=90°, and spatial resolution=0.78x0.78x0.78mm. Preprocessing. T2-weighted MRI images were corrected for N4 inhomogeneity field4 and aligned to the MNI template using non-linear registration and Advanced Normalization Tools5. Algorithm. The proposed PSD segmentation method is based on a combination of Step-A: extraction of a parasagittal space mask using a U-Net model6 with 3D patches as inputs (96x64x64voxels) followed by a parasagittal probabilistic atlas built from the mean of manual segmentations. Step-B: labelling of PSD voxels using a bi-modal Gaussian mixture model, an optimal threshold is computed with a maximum a posteriori and expectation-maximization algorithm (Figure 1). Evaluation. PSD space from all participants was manually delineated, by a board-certified neuroradiologist (experience=12 years), along the superior sagittal sinus extending from its most anterior aspect along the frontal calvarium to the region along the torcula posteriorly. We evaluated the accuracy using Sørensen–Dice coefficient (DSC), true positive (TPF), false positive (FPF), and false negative fraction (FNF) in a leave-one-out cross-validation procedure (14 images used for training, 4 for validation, and 1 for testing). One-way ANOVA was used to assess the differences between each group.Results
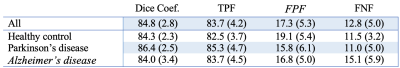
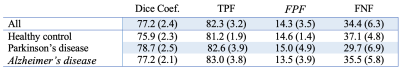
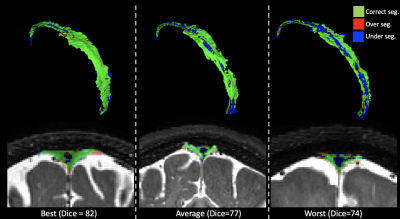
Method performance was evaluated for each step. Step A. Table 2 summarizes the segmentation accuracy of the extraction of PSD space. Performance assessment of all subjects obtained a DSC of 84.8, TPF of 83.7%, FPF of 17.3%, and FNF of 12.8%. Importantly, no significant differences were observed between groups, highlighting algorithm generalizability. Step B. Table 3 summarizes the accuracy of the PSD volume segmentation using automatic voxel labeling. Accuracy assessment of all participants obtained a DSC of 77.2, TPF of 82.3%, FPF of 14.3, and an FNF of 34.4%. Similarly, as above, no significant differences were observed for the segmentation of each individual group. Examples of correct, under, and over-segmentation for the best, an average, and the worst segmentation maps are provided in Figure 2.Discussions
To our knowledge, this is the first method proposed to extract the PSD space using non-contrast MR imaging. Importantly, our method only requires T2-weighted MRI to accurately extract the volume of the region located alongside the sagittal sinus, which has recently been postulated to contain cerebral lymphatic channels. The minimal pre-processing steps and the compacity of our model enable the method to be robust to anatomical variability secondary to neurodegeneration. Moreover, Table 2 shows the comparison between the automatic segmentation and delineation of sinus veins and PSD space using venous blood flow contrast. Venous structures and dura appear hypointense on the T2-weighted MRI relative to the PSD space, enabling the use of an intensity-based threshold method to delineate PSD voxels from the rest of the parasagittal space mask computed by using deep learning. This suggests that the proposed method can be effectively applied to quantify the PSD, even without additional source imaging or angiography. These findings should greatly increase abilities to evaluate PSD space even using historical datasets, which should provide an important resource for future investigations of possible glymphatic dysfunction.Conclusions
We provide evidence that automatic segmentation of the PSD space can be accurately achieved using non-contrast 3D T2-weighted MRI only. This procedure incorporates a novel method based on a combination of deep-learning and T2-weighted intensity thresholding. The method yields comparable accuracy of PSD extraction as methods that require exogenous contrast and supplementary acquisitions of venography. Ongoing work is focused on evaluating the generalization of different 3D T2-weighted MRI sequences, as well as the physiological relevance of these findings in the growing number of applications where glymphatic dysfunction is implicated.Acknowledgements
This study was supported by NIH/NIA 5R01AG062574 and NIH/NCCIH 1R01AT011456.References
1. Iliff, J. J. et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 4, (2012).
2. Ringstad, G. & Eide, P. K. Cerebrospinal fluid tracer efflux to parasagittal dura in humans. Nat. Commun. 11, (2020).
3. Park, M., Kim, J. W., Ahn, S. J., Cha, Y. J. & Suh, S. H. Aging Is Positively Associated with Peri-Sinus Lymphatic Space Volume: Assessment Using 3T Black-Blood MRI. J. Clin. Med. 2020, Vol. 9, Page 3353 9, 3353 (2020).
4. Tustison, N. J. et al. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010).
5. Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011).
6. Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 9351, 234–241 (2015).
Figures