2224
Hippocampal Subfields Volume in Middle Age Healthy Adults
salem Alkhateeb1, Tales Santini1, Li jinghang1, Robin Chu1, Daniel Ibrahim1, Anna M. Marsland1, Stephen B. Manuck1, Pete Gianaros1, and tamer S. Ibrahim2
1University of Pittsburgh, Pittsburgh, PA, United States, 2University of Pittsburgh, pittsburgh, PA, United States
1University of Pittsburgh, Pittsburgh, PA, United States, 2University of Pittsburgh, pittsburgh, PA, United States
Synopsis
As hippocampal volume has been extensively utilized as a diagnosing tool to confirm diagnosis of many neurological disorders, this study aims to employ the high resolution 7T TSE T2w data to segment high quality images with precision based on multi atlases and machine learning. Results have proved that this pipeline provides excellent outcomes and is validated to be used for more variables.
Introduction
The hippocampus (HC) plays an important role in retaining and working memories, learning processes and attention (1). The atrophy of the hippocampus system and connectivity are associated with several neurological disorders and normal aging (2). Hippocampus structure has been well studied and the hippocampal level of atrophy and cognitive decline in healthy population are related to age and environmental factors such as hardships (3). Therefore, quantifying hippocampus volume precisely has become a main variable in the analysis of a targeted population, including the ones with high risk of dementia and Alzheimer’s disease. The recent hardware and software advancements in 7T MRI scanner technology have shown unprecedent higher spatial resolution and contrast images enabling higher anatomical visualization of the hippocampal subfields that are divided into cornu ammonis fields (CA1, CA2, CA3), the dentate gyrus (DG), the entorhinal area (EC), and the subiculum (Sub) (4). Segmenting the hippocampal subfields allows researchers to longitudinally and cross-sectionally extend the collective knowledge of the hippocampal subfield’s functionality. However, hippocampus segmentation at 7T MRI is not trivial and requires rigorous preprocessing, highly trained raters, and consistency between multi centers. Therefore, the emerging multiple atlas based automated segmentation of hippocampal subfields (ASHS) package has gained the attention of multi center research labs for the excellent outcomes and reproducibility provided (7). The aim of this study is to perform a preliminary group statistical analysis on 226 healthy midlife adults, the outcome of this work is to validate the hippocampus segmentation pipeline implemented using ASHS package but tailored to our 7T MRI system and protocols. The expectation of hippocampal subfields volumes shrinkage with age is used as an indicator to assess the outcomes of the pipeline that will be utilized to associate the levels of inflammation markers and hippocampal sub regional volumes.Methods
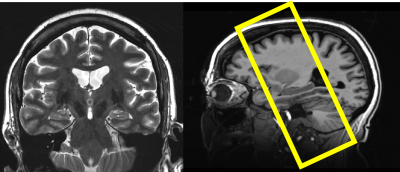
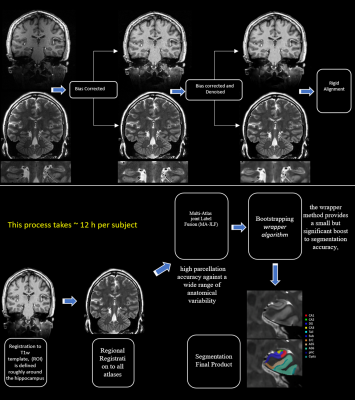
All images were collected using a 7T MRI scanner (Siemens Magnetom, Germany) and a 16Tx/32Rx Tic-Tac-Toe (TTT) RF head coil system, the performance of this design has been validated in more than 2000[TS1] [GU2] participant scans which are part of numerous neuroimaging studies at University of Pittsburgh. 0.75 mm isotropic T1-weighted (T1w) Magnetization Prepared RApid Gradient Echo (MPRAGE) acquisition and (0.375 x 0.375 x 1.5 mm3) T2-weighted slanted (perpendicular to the main hippocampal axis) 2D turbo spin echo (TSE) as shown in figure (1).In collaboration with ‘Community socioeconomic disadvantage in midlife relates to cortical morphology via neuroendocrine and cardiometabolic pathways’ research study group, data from 226 participants (age 57 ± 7.6 years old) were collected for this work. Segmentation was collected using the pipeline illustrated in the flowchart in figure (2). An additional final step is necessary to manually correct subfields that are mislabeled, most commonly in images that include motion, this quality assurance step is achieved by following the slice-by-slice guidelines from literature to contain reproducibility (5).
Results
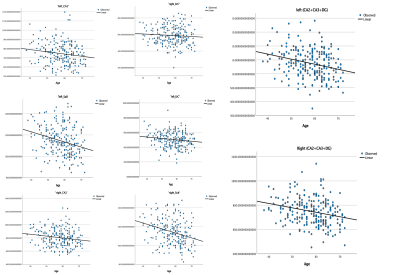
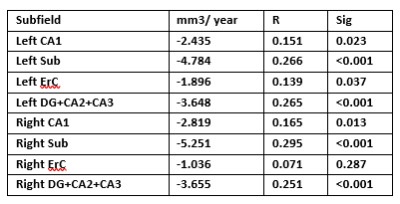
Linear regression of the hippocampal subfields volumes in correlation with age are illustrated in figure (3) and ANOVA output for each subfield is described in table (1). We observed a reduction in volume of each subfield in both hemispheres, and a significant difference was observed in all subfields, except the Right ErC, which presented a trend that indicates association between atrophy and age.Discussion and Conclusion
In this preliminary group analysis, we acquired high –resolution and high SNR images (MPRAGE T1w, TSE T2w) from 226 subjects at 7T, enabling a good visualization of the hippocampal subfields to use them as our input to a pipeline we implemented using the ASHS package with modifications and additions to optimize it to our own protocols and systems. The images were used to validate our pipeline output and quality check the grade of the final segmentation. After performing statistical analysis on this group, we found that hippocampal subfields shrinkage is highly correlated with age, as we expected. However, one subfield Right ErC Left showed a non-significant association with age, but we observed a trend in volume reduction. The findings show that our atlas-based fully automated pipeline and post-processing stage provide a highly anatomical labeling of each subfield. Future work will use this segmentation results to correlate the hippocampus subfields volumes with inflammation markers, after adjusting for intracranial volume, age, and sex.Acknowledgements
This work was supported by the National Institutes of Health under award numbers R01DK110041, R01MH111265, R01AG056043, R01AG063525,and T32MH119168 And in part by the University of Pittsburgh Center for Research Computing through the resources provided.References
1. Eichenbaum H. The hippocampus and declarative memory: cognitive mechanisms and neural codes. Behav Brain Res. 2001 Dec;127(1–2):199–207.2. Lisman J, Buzsáki G, Eichenbaum H, Nadel L, Ranganath C, Redish AD. Viewpoints: how the hippocampus contributes to memory, navigation and cognition. Nat Neurosci. 2017 Nov 1;20(11):1434–47.3. Gianaros PJ, Kuan DC-H, Marsland AL, Sheu LK, Hackman DA, Miller KG, et al. Community Socioeconomic Disadvantage in Midlife Relates to Cortical Morphology via Neuroendocrine and Cardiometabolic Pathways. Cereb Cortex. 2015 Oct 22;bhv233.4. Giuliano A, Donatelli G, Cosottini M, Tosetti M, Retico A, Fantacci ME. Hippocampal subfields at ultra high field MRI: An overview of segmentation and measurement methods: SEGMENTATION AND MEASUREMENT METHODS FOR HIPPOCAMPAL SUBFIELDS. Hippocampus. 2017 May;27(5):481–94.5. Hasselmo ME. The Role of Hippocampal Regions CA3 and CA1 in Matching Entorhinal Input With Retrieval of Associations Between Objects and Context: Theoretical Comment on Lee et al. (2005). Behav Neurosci. 2005;119(1):342–5.6. Eichenbaum H. The hippocampus and declarative memory: cognitive mechanisms and neural codes. Behav Brain Res. 2001 Dec;127(1–2):199–207.7. Yushkevich PA, Pluta JB, Wang H, Xie L, Ding S-L, Gertje EC, et al. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment: Automatic Morphometry of MTL Subfields in MCI. Hum Brain Mapp. 2015 Jan;36(1):258–87.8. Berron D, Vieweg P, Hochkeppler A, Pluta JB, Ding S-L, Maass A, et al. A protocol for manual segmentation of medial temporal lobe subregions in 7 Tesla MRI. NeuroImage Clin. 2017;15:466–82
DOI: https://doi.org/10.58530/2022/2224