2223
Lung segmentation with deep learning for 3D MR spirometry1Université Paris-Saclay, CEA, CNRS, Inserm, BioMaps, Orsay, France, 2IADI U1254, INSERM, Université de Lorraine, Nancy, France
Synopsis
Current MR lung image segmentation has huge challenges compared to CT images, specifically in terms of low contrast, non-homogeneity. We developed a series of processing to accelerate the manual correction of reference standard lung masks by the auto-seeds region growing. Finally, we developed a 3D automatic MRI lung segmentation method using deep learning with a limited dataset (41 volumes for training and 4 volumes for validation). In the primary result, this lung segmentation archived a Dice score of 0.917±0.013. In the case of limited data, it provides us a new way for MR lung segmentation.
INTRODUCTION
The regional quantification of lung function by MRI requires determining the organ boundaries. However, while successful for X-ray computed tomography, automated lung segmentation usually fails for MRI. Therefore, in the framework of 3D Magnetic Resonance Spirometry [1] for assessing lung function on a regional scale, we developed an automatic segmentation method using deep learning with a limited dataset.METHODS
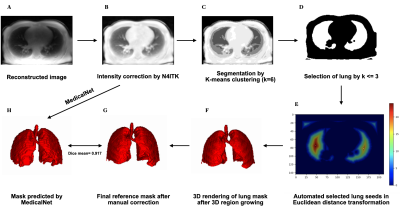
Image Acquisition Protocol: Three-dimensional dynamic lung images were acquired in 21 healthy human volunteers spontaneously breathing over a 10-minute scan while lying in four positions (supine, prone, decubitus left and right) in a GE Signa PET/MR 3.0 T machine using a UTE pulse sequence (TE = 14 μs, TR = 2 ms, flip angle = 3°, Voxel size = 1.5 mm isotropic, Matrix size = (212×212×142) voxels, Bandwidth =100 kHz, RF spoiling) along optimal AZTEK trajectories [2].Reference Standard Lung Segmentation: Dataset were preprocessed with (1) intensity correction using N4ITK algorithm [3] (Figure 1 B), (2) pre-segmentation on the corrected images by auto-seed region growing [4] (Figure 1 C D E F) and (3) manual correction to obtain a reference standard lung mask (Figure 1 G). Under the supervision of a medical doctor, we finally completed the manual segmentation of 21 patients with a total of 45 supine volumes.
Dataset: The dataset consisted of 45 intensity corrected volumes as input and their corresponding masks as output. Forty-one volumes were used for training and 4 volumes, for validation. All the data was normalized and resized to the same size (150,200,200).
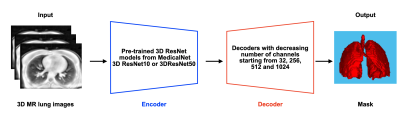
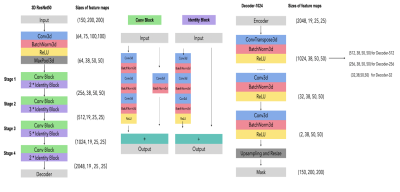
Deep Learning Encoder-Decoder Architecture: The MedicalNet project aggregated the datasets from several medical challenges to build a relatively large dataset across diverse modalities (including CT and MRI), target organs (including the lungs) and diseases [5]. Due to the lack of great quality and quantity of MR lung datasets to learn from scratch, our neural network structure consisted of a pre-trained model of 3D ResNet from MedicalNet, as the encoder, and a decoder of three groups layers. The first set of decoder layers comprised a transposed convolution layer with a kernel size of (3×3×3) and 512 channels, which was used to amplify twice the feature map (up-sampling). These were followed by several convolutional layers with (3×3×3) kernel size with numbers of channels reduced by half in turn progressively until 32. Finally, the convolution layer in the (1×1×1) kernel was employed to generate the final output. The number of channels corresponds to the number of categories (Figure 2).
Materials: GPU: Tesla k40 c, Python: 3.7.0 and Pytorch: 1.2.0
Training: The network parameters were optimized using the cross-entropy loss with the standard stochastic gradient descent (SGD) method for which the learning rate was set to 0.1, the momentum, to 0.9 and the weight decay, to 0.01 [5]. Additionally, the learning rate decayed exponentially by a coefficient of 0.99 with the epoch to accelerate the learning in the early epochs and slow down the learning in the later epochs so more detailed features could be obtained.
RESULTS
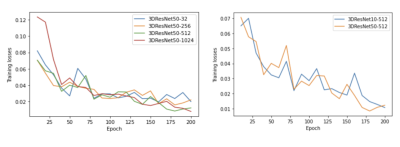
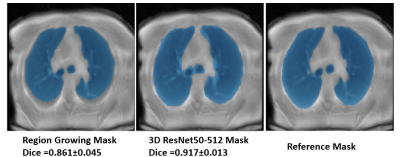
The N4ITK algorithm was proven to be robust for the intensity correction of lung MR images (Figure 1 B). The auto-seeds region growing method achieves a Dice score of 0.861±0.045 (Figure 5 left). The best network structure is the 3D ResNet50-512 with a Dice score of 0.917±0.013 (Figure 5 center) and a minimal training loss value of 0.01 (Figure 4).DISCUSSIONS
(1) Compared with Zha et al. [4], our pre-segmentation method using auto-seeds region growing was strongly overperformed. (2) The single metric Dice coefficient does not provide all relevant information to properly evaluate segmentation. This should be assessed using multiple criteria. (3)The model may encounter the problem of over-fitting and not finding the global minimum. (4) Better segmentation results may be achieved by using a progressive up-sampling decoder.CONCLUSIONS
The newly developed automatic segmentation method using deep learning is successful. It greatly saves time from manual segmentation while providing high accuracy. The model could be further improved by changing the more advanced optimizer, augmenting the dataset, optimizing the learning rate and using a progressive up-sampling decoder. Finally, the neural work 3D ResNet50-512 could be used for deep learning multi-label segmentation to get the parenchyma, the vascular tree and the large airways.Acknowledgements
The PET/MR platform is affiliated with the France Life Imaging network (grant ANR-11-INBS-0006).References
[1] T. Boucneau, B. Fernandez, P. Larson, L. Darrasse, and X. Maître, “3d magnetic resonance spirometry,”Scientific Reports, vol. 10, 12 2020.
[2] T. Boucneau, B. Fernandez, F. L. Besson, A. Menini, F. Wiesinger, E. Durand, C. Caramella, L. Darrasse, and X. Maître, “Aztek: Adaptsive zero te k-space trajectories,” Magnetic Resonance in Medicine, vol. 85, pp. 926–935, 2 2021.
[3] N. J. Tustison, B. B. Avants, P. A. Cook, Y. Zheng, A. Egan, P. A. Yushkevich, and J. C.Gee, “N4itk: Improved n3 bias correction,” IEEE Transactions on Medical Imaging, vol. 29, pp. 1310–1320, 6 2010.
[4] W. Zha, S. B. Fain, M. L. Schiebler, M. D. Evans, S. K. Nagle, and F. Liu, “Deep convolutional neural networks with multiplane consensus labeling for lung function quantification using ute proton mri,” Journal of Magnetic Resonance Imaging, vol. 50, pp. 1169–1181, 10 2019.
[5] S. Chen, K. Ma, and Y. Zheng, “Med3d: Transfer learning for 3d medical image analysis,” 4 2019. Pre-trained models and relative source code has been made publicly available at https://github.com/Tencent/MedicalNet.
Figures