2164
A deep-learning non-contrast MRI lesion segmentation model for liver cancer patients that may have had previous surgery1Perspectum, Oxford, United Kingdom
Synopsis
Segmentation of lesions within the liver is pivotal for surveillance, diagnosis and treatment planning, and automated or semi-automated approaches can aid clinical workflows. Many patients that are under surveillance may have had previous surgery, meaning their scans will contain post-surgical features. We investigate the use of a deep learning segmentation model for non-contrast MRI that can distinguish between lesions, surgical clips and resection-induced fluid-filling regions. We report mean dice scores of 0.55, 0.72 and 0.88 for these classes respectively, demonstrating the potential of this model for semi-automated workflows.
Introduction
Liver cancer is one of the leading causes of death worldwide and is on the rise1. Localisation and segmentation of lesions within the liver is a prerequisite for diagnosis and treatment planning, but manual segmentation is time consuming and prone to variability. Therefore, automated or semi-automated approaches can greatly aid clinical workflows2.The fact that many patients have had surgery for a previous tumour3, and thus show features such as surgical clips or fluid accumulations in their scans, impedes automated assessments (e.g. liver and lesion volume assessments for treatment planning). The ability to segment these features and distinguish them is therefore critical, and models such as these have not been investigated to such a great extent in literature. Furthermore, many studies about automatic lesion segmentation focus on CT and/or contrast-enhanced images4. Studies making use of non-contrast MRI data are less common. However, abbreviated protocols with non-contrast MRI are becoming more popular5 due to lower cost, higher throughput, and the fact that contrast agents are not suitable for all patients.
In this study, we investigate a deep learning U-Net model for segmenting lesions, surgical clips and fluid-filled regions from non-contrast MRI data in a dataset that contains both pre- and post-surgical cases. We compare the feasibility of segmenting each of the classes and compare models for optimal class groupings. To our knowledge, this is the first study to have investigated a liver lesion segmentation model targeted to non-contrast MRI scans for cases with previous surgery.
Methods
The dataset consisted of 200 subjects, with some subjects scanned at multiple time-points, resulting in a total of 343 volumetric T1-weighted scans. The scan annotations had the following four labels: liver parenchyma (including vessels), lesions (including primary cancer and metastases), surgical clips and fluid-filled regions. Examples of these classes are shown in figure 1. The scans were split into 141 subjects (243 scans) training, 19 (34) validation and 40 (66) test. Note that we are planning on using the test set for a validation of the final chosen model once this study is completed, hence the results shown here are on the validation set only.We compared two models: one for the segmentation of 4 target classes (background liver, lesion, surgical clips and fluid-filled/other), and another for 2 target classes, grouping classes 2, 3 and 4 to a joint abnormality class.
The 3D U-Net architecture for the semantic segmentation model is show in figure 2, which is based on the work in6. To input images into the model, the pre-processing steps included cropping around the liver region by first applying a separate low-resolution liver delineation model, clamping and normalising intensities, resampling the input image to a 1.5x1.5x3 mm resolution, and extracting patches of size 192x192x64. Models were trained until the loss on the validation set stopped improving.
Results
Figure 3 shows the dice scores on the validation set for the 4-class and 2-class models. Note that if both the ground truth and predicted masks were empty for a given class, then a dice score of 1 was assigned for that sample. Figure 4 (top) shows an example where the models performed well, while figure 4 (bottom) shows one example where the models struggled with under-segmentation.Discussion
We have developed a model that can be applied to cases that may have had previous surgery, which enables the segmentation of other abnormalities in addition to typical lesions, such as surgical clips and fluid-filled regions in non-contrast MRI.In general, the dice scores and our qualitative assessment of the segmentations are very promising, especially given the size of the dataset, the fact that we process non-contrast data and that many surgical clips and lesions are small. This means that there is scope for the exploration of the proposed models in semi-automated pipelines, for example where an operator manually checks and edits the segmentations.
We observed that a simplification to 2 classes did not improve the performance for cases in which the 4-class model performed inaccurately and found that overall, the 2-class model was likely to result in more false positives. Given that the performances were not substantially different, the 4-class model would be the preferable choice, since it gives a more fine-grained separation of the classes.
For future work, we will investigate techniques for addressing the type of under segmentation that can be seen in the bottom of figure 4. This may have been caused by class imbalance and could be tackled through more appropriate cost functions. In addition, a two-stage approach that is more sensitive at the first stage and refines the predictions in the second stage could be considered.
Conclusion
This study investigated developing the use of a deep learning segmentation model targeted towards applications such as treatment planning or surveillance, where subjects that may have had previous surgery. We showed that it is feasible to develop such a model for use with non-contrast MRI and that there is scope for its use in semi-automated pipelines. We also showed that using a 4-class model is preferable to a coarse 2-class model.Acknowledgements
No acknowledgement found.References
1. Ryerson A.B., et al. Annual Report to the Nation on the Status of Cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer. 2016, 122(9):1312-37.
2. Mojtahed A., et al. Repeatability and reproducibility of deep-learning-based liver volume and Couinaud segment volume measurement tool. Abdom Radiol. 2021 Epub ahead of print. doi: 10.1007/s00261-021-03262-x
3. Hyder O., et al. Post-treatment surveillance of patients with colorectal cancer with surgically treated liver metastases. Surgery. 2013, 154(2):256-65.
4. Bilic, P. et al. The liver tumor segmentation benchmark (lits). 2019, arXiv preprint arXiv:1901.04056.
5. Khatri G., et al. Abbreviated-protocol screening MRI vs. complete-protocol diagnostic MRI for detection of hepatocellular carcinoma in patients with cirrhosis: An equivalence study using LI-RADS v2018. J Magn Reson Imaging. 2020, 51(2):415-425.
6. Arya Z., et al. Deep learning-based landmark localisation in the liver for Couinaud segmentation. Medical Image Understanding and Analysis, pp.227-237, Springer, Cham 2021.
Figures
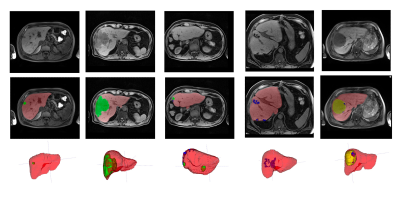
Figure 1: Examples of the classes in the dataset. Left to right: small lesion pre-resection, large lesion pre-resection, multi-focal lesion after resection, surgical clips after resection, fluid filling after resection.
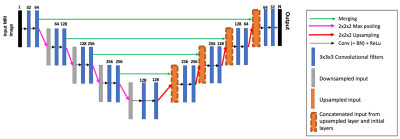
Figure 2: The 3D U-Net architecture used for semantic segmentation. There is a softmax activation on the output and N is the number of class labels (the background was included as its own class).
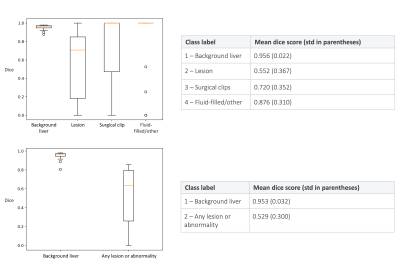
Figure 3: Top – the dice scores for the 4-class model on the validation set. Bottom – the dices scores for the 2-class model.
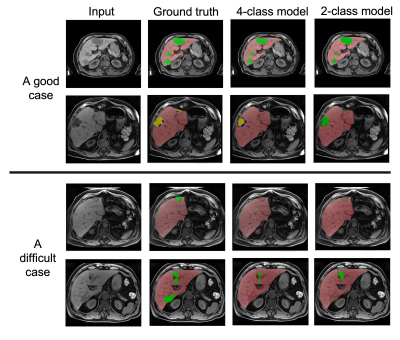
Figure 4: The input image, ground truth segmentation, 4-class model prediction and 2-class prediction for (top) a good case and (bottom) a difficult case.