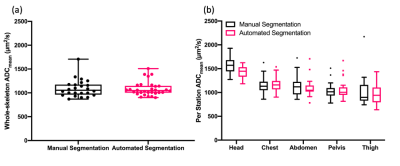2162
Automatic segmentation of whole-body MRI using UnnU-Net: Feasibility of whole-skeleton ADC evaluation in plasma cell disorders1Department of Biomedical Engineering and Imaging Science, King's College London, London, United Kingdom, 2School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom, 3University College London, London, United Kingdom, 4Department of Radiology, Guy’s & St Thomas’ NHS Foundation Trust, London, United Kingdom, 5Siemens Healthcare Limited, London, United Kingdom
Synopsis
Multiple myeloma is a heterogeneous bone marrow cancer. Assessment of changes in mean apparent diffusion coefficient (ADCmean) is helpful for evaluating treatment response but has not been feasible for the whole skeleton due to the time-consuming nature of manual segmentation. Whole-skeleton and per-station ADCmean were quantified from whole-body MRI using automated segmentation by an uncertainty-aware nnU-Net in 30 patients with plasma cell disorders and compared against the manual segmentation by radiologists. No differences were observed in whole-skeleton or per-station ADCmean when using the automatic and manual segmentations. Further investigation is required in a larger dataset, but initial results are promising.
Introduction
Multiple myeloma (MM) is a bone marrow cancer characterised by clonal plasma cells (1). 140,000 new cases are diagnosed each year with 106,000 deaths (2). Whole-body MRI (WB-MRI) is a sensitive modality for detecting both focal myeloma lesions and diffuse disease (3). Currently, the presence of at least one MRI-detected focal lesion is a myeloma-defining event (4), however, marrow infiltration may be evident on MRI prior to the bone destruction causing focal lesions, and disease may be heterogeneously distributed (5). Nowadays, the change in the mean value of apparent diffusion coefficient (ADC) following treatment, computed on a region-of-interest (ROI) placed within a lesion, is used to assess treatment response (6). However, limited ROI analysis of a diffuse pattern of disease may be unrepresentative. Therefore, the mean ADC (ADCmean) computed over the whole skeleton may be a better indicator of treatment response, but the manual segmentation of the skeleton on WB-MRI is time-consuming for a radiologist. In this abstract, we assess whether the automatic uncertainty-aware nnU-Net (UnnU-Net) that we proposed previously (7) can be used to reproduce the whole-skeleton ADCmean computed by a radiologist.Methods
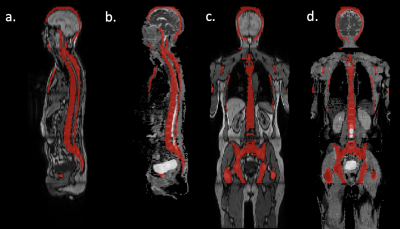
Patients with suspected plasma cell disorders underwent 1.5T WB-MRI (Magnetom Aera, Siemens) from skull vertex to thigh, acquired in 5 stations. Axial T1-weighted Dixon gradient-recalled echo (GRE) (TR=6.62 ms; TE=4.77, 2.39 ms; flip angle 10°; NEX=1; FOV 500 mm; reconstructed matrix 640, reconstructed voxels=0.8x0.8 mm, reconstructed slice thickness=5 mm) and axial diffusion-weighted echo-planar sequences (TR=6270 ms; TE=67 ms; flip angle 90°; NEX=1; b-values: 50, 900 s/mm2; FOV 500 mm; reconstructed matrix 256, reconstructed voxels=2.0x2.0 mm, reconstructed slice thickness=5 mm) were acquired.Our UnnU-Net segmentation algorithm (7) was applied to the axial per-station T1-weighted Dixon GRE images. An example of automated segmentation is shown in Figure 1. Automated segmentations from each station were resampled and padded to ensure the same spatial size and pixel resolution as ADC maps.
The per-station and whole-skeleton ADCmean values derived from UnnU-Net segmentation were compared to those derived from manual segmentation at the patient level. The comparison was statistically evaluated using the Wilcoxon signed-rank test at 5% significance.
Results
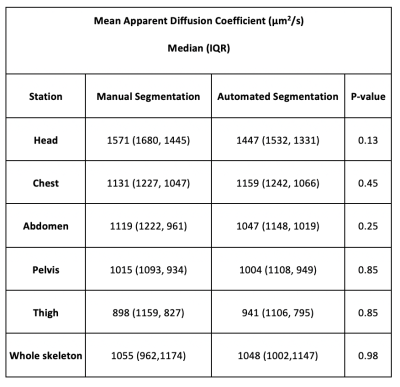
Thirty patients were included in the study. Table 1 summarises the median (IQR) ADCmean for the different stations and whole skeleton computed over 30 patients. Differences between the distributions of both whole-skeleton and per-station ADCmean derived using manual and automated segmentations were not significant (Table 1). Figure 2 shows the box and whisker plots of ADCmean of manual and automated segmentations.Discussion
Multiple myeloma is known to be a heterogeneous disease with different patterns of involvement within the skeleton (3,7). There is an ongoing clinical need for a more objective measure of disease extent throughout the skeleton e.g., for assessing treatment response. Manual segmentation of the whole skeleton is time-consuming and not viable for clinical practice. We have shown that UnnU-Net segmentation of the whole skeleton is feasible with minor errors and that there are no significant differences between the whole-skeleton and per-station ADCmean values from manual and UnnU-Net segmentation.The subjects in this study reflected the spectrum of plasma cell disorders, and ADC values were broadly consistent with the literature (viable tumour range of 700-1400 µm2/s, with higher values for treated tumours) (5). Per-station analysis also showed that the ADCmean decreased from the head to thigh stations for both manual and automated segmentations; this in part reflects the underlying age-related red-yellow marrow distribution but also difficulties in segmenting the skull both manually and automatically.
Conclusion
ADCmean computed using automated segmentation of our algorithm is not statistically different to ADCmean computed using manual segmentation. This is a step forward for objective assessment of disease extent, complementing focal lesion assessment in treatment-response. Further investigation of whole-skeleton ADC as a biomarker is needed.Acknowledgements
This work was supported by the EPSRC Centre for Doctoral Training in Medical Imaging (EP/L015226/1).References
1. Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. American Journal of Hematology 2020;95:548–567 doi: 10.1002/ajh.25791.
2. Cowan AJ, Allen C, Barac A, et al. Global burden of multiple myeloma: A systematic analysis for the global burden of disease study 2016. In: JAMA Oncology. Vol. 4. American Medical Association; 2018. pp. 1221–1227. doi: 10.1001/jamaoncol.2018.2128.
3. Dimopoulos MA, Hillengass J, Usmani S, et al. Role of Magnetic Resonance Imaging in the Management of Patients With Multiple Myeloma: A Consensus Statement. Journal of Clinical Oncology 2015;33:657–664 doi: 10.1200/JCO.2014.57.9961.
4. Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. The Lancet Oncology 2014;15:e538–e548 doi: 10.1016/S1470-2045(14)70442-5.
5. Messiou C, Hillengass J, Delorme S, et al. Guidelines for Acquisition, Interpretation, and Reporting of Whole-Body MRI in Myeloma: Myeloma Response Assessment and Diagnosis System (MY-RADS). Radiology 2019;291:5–13 doi: 10.1148/radiol.2019181949.
6. Messiou C, Giles S, Collins DJ, et al. Assessing response of myeloma bone disease with diffusion-weighted MRI. http://dx.doi.org/10.1259/bjr/52759767 2014;85 doi: 10.1259/BJR/52759767.
7. Gu R, Antonelli M, Mehta P, Amlani A, Green A, Neji R, Ourselin S, Dregely I, Goh V. Using uncertainty estimation to increase the robustness of bone marrow segmentation in T1-weighted Dixon MRI for multiple myeloma. In: ISMRM 2020 Poster Presentation, Charlotte, United States; 2020. https://www.ismrm.org/21/program-files/TeaserSlides/TeasersPresentations/2409-Teaser.html
Figures