2154
Incorporating Histological Grading for Brain Tumor Segmentation1The Centre of Mathematical Imaging in Healthcare, Department of Applied Mathematics and Theoretical Physics, University of Cambridge, Cambridge, United Kingdom, 2Cambridge Brain Tumor Imaging Laboratory, Division of Neurosurgery, Deoartment of Clinic Neuroscience, Univerisity of Cambridge, Cambridge, United Kingdom
Synopsis
Automatic tumor segmentation on MRI is crucial for patient management of brain tumors. However, previous automatic MRI segmentation methods based on deep learning are limited by bottlenecks of feature extraction. In this paper, we seek to investigate the influence of incorporating prior histology grading on the performance of tumor segmentation. We propose to employ histological grade labels to strengthen the feature extraction and tumor localization in the encoder, which can boost the DSC at exceeding 2% in different U-Net baselines. Our results could support the usefulness of incorporating clinical prior knowledge to improve the segmentation performance without introducing extra parameters.
Introduction
Glioblastoma is the most common primary brain cancer, characterized by remarkable tumor heterogeneity and infiltrative growth [1,2]. According to the World Health Organization (WHO), glioma can be classified into four grades, where lower-grade glioma (LGG) includes grade II and grade III [3]. Magnetic resonance imaging (MRI) provides a non-invasive approach for tumor diagnosis and characterization [4]. Based on MRI, a precise tumor segmentation is crucial for treatment planning and longitudinal follow-up. However, manual segmentation is often laborious and prone to a high inter-observer variability [5].Recently, deep learning-based methods have achieved excellent performance in tumor segmentation by extracting non-hand-crafted features. However, the current frameworks have encountered the bottleneck in performance due to the limitation of computational resources. On the other hand, for natural image classification, prior knowledge, such as pre-trained model [6] and multi-task head [7], has been shown to boost the model performance. Therefore, incorporating clinical prior knowledge shows potential to improve the segmentation performance in brain tumors. In this work, we investigate the performance of including histological grading as the prior knowledge without introducing extra tuning parameters. Our results show that the classification of tumor grading facilitates any encoder backbone by extracting more relevant imaging features for tumor segmentation.
Methods
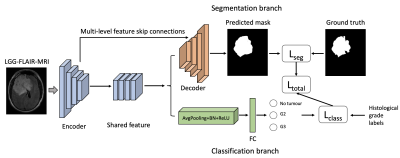
The overall study design is shown in Figure 1. The proposed method consists of an encoder and multi-task branches. The shared feature maps from the bottom stage of the encoder are fed into the segmentation branch and classification branch. The multi-level feature skip connections from the U-Net are preserved in the segmentation branch. In the classification branch, the region beyond the tumor on FLAIR MRIs is labelled as no tumor. The images with tumors are classified into grade II (G2) or grade III (G3). The histological grading can promote feature extraction and tumor localization in the encoder. This model design could maintain the amounts of tuning parameters in segmentation and inference time in testing. In the training process, we employed the sum of binary cross entropy and dice as the segmentation loss in the segmentation section for the consideration of faster convergence and optimal segmentation region. For classification heads, a cross entropy loss with label smoothing is used for better generalization. The total loss is shown below functions. The multi-task will be optimized simultaneously and benefit each other.$$$L_{total}=-\frac{1}{N}\sum_{i=1}^{N}{((y\ log(x)\ +\ (1\ -\ y)\ log(1\ -\ x))}+2\frac{X\cap Y}{\left|X\right|+\left|Y\right|}+\left(-\sum_{c=1}^{M}{p_c\log{\left(q_c\right)}}\right)$$$
$$$q_c= \begin{cases} 1- \alpha& \text{if c=y,}\\ \frac{\alpha}{1-M}& \text{otherwise} \end{cases}$$$
Here, $$$N$$$ denotes the pixel number in mask $$$y$$$ and predicted mask $$$x$$$. $$$M$$$ represent the number of categories. The $$$p$$$ and $$$q$$$ are label and predicted probability. $$$\alpha$$$ is a constant for label smoothing.
Results
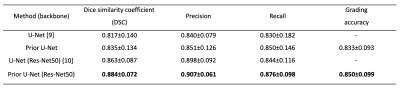
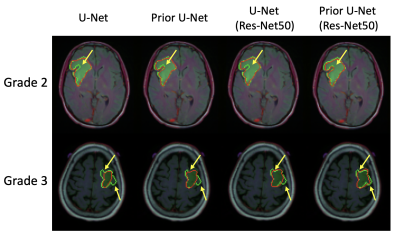
A total of 109 patients with G2 and G3 gliomas were included from The Cancer Genome Atlas (TCGA) lower-grade glioma collection (TCGA-LGG) [8]. We split the patients into 77 patients (2728 images) for training and 16 patients (574 images) for validation, and another 16 patients (589 images) for testing.Example predictions and ground truth visualizations are shown in Fig. 2. The U-Net and Res-Net50 U-Net with prior grade classification show that the prediction (red line) is closer to ground truth (green line) than original structures. The prior U-Net with Res-Net50 backbone achieves the highest coincidence with dice similarity coefficient (DSC) of 0.884, as shown in Table 1. In addition, the performance of vanilla U-Net is also improved after introducing the prior grade classification. The classification branch obtain more than 83% accuracy.
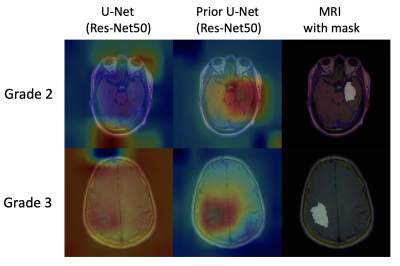
We conducted the classification activation maps (CAMs) [11] approach from the U-Net (Res-Net50) and the prior U-Net (Res-Net50) to identify the potential reason for performance improvement. As shown in Fig. 3,, the prior grading information can help the models obtain a more concentrated attention on relevant tumor regions, suggesting more accurate activation regions could promote better segmentation performance in decoding.
Conclusion
This work illustrates that the prior information from histology grading can assist deep learning models to focus on tumour region and provide more efficient automatic tumour segmentation without increasing parameter space. Meanwhile, this multi-tasking pipeline including segmentation and grading branches allows for fully automatic and simultaneous segmentation and grade classification of brain tumours. Additionally, we observe a segmentation DSC improvement of more than 2%, increased precision and recall rate, and tumour grading accuracy at image and patient levels of exceeding 83%, respectively, which supports that effective prior histology knowledge promises to act as a feasible tool for tumour characterization. In future work, we will explore the possibilities of merging prior histological information with multi-contrast brain MRI for segmentation by developing semi- or weakly supervised learning approaches.Acknowledgements
.References
1. Zadeh Shirazi, A., McDonnell, M.D., Fornaciari, E. et al. A deep convolutional neural network for segmentation of whole-slide pathology images identifies novel tumour cell-perivascular niche interactions that are associated with poor survival in glioblastoma. Br J Cancer 125, 337–350 (2021). https://doi.org/10.1038/s41416-021-01394-x
2 . Louis, D.N., Ohgaki, H., Wiestler, O.D. et al. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol 114, 97–109 (2007). https://doi.org/10.1007/s00401-007-0243-4
3. Louis, D.N., Perry, A., Reifenberger, G. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131, 803–820 (2016). https://doi.org/10.1007/s00401-016-1545-1
4. Mohamed A. Naser, M. Jamal Deen. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Computers in Biology and Medicine, Volume 121, 2020, 103758, ISSN 0010-4825, https://doi.org/10.1016/j.compbiomed.2020.103758.
5. Ranjbarzadeh, R., Bagherian Kasgari, A., Jafarzadeh Ghoushchi, S. et al. Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci Rep 11, 10930 (2021). https://doi.org/10.1038/s41598-021-90428-8
6. He, K., Girshick, R.B., & Dollár, P. (2019). Rethinking ImageNet Pre-Training. 2019 IEEE/CVF International Conference on Computer Vision (ICCV), 4917-4926.
7. Crawshaw, M. (2020). Multi-Task Learning with Deep Neural Networks: A Survey. ArXiv, abs/2009.09796.
8. Pedano, N., Flanders, A. E., Scarpace, L., Mikkelsen, T., Eschbacher, J. M., Hermes, B., … Ostrom, Q. (2016). Radiology Data from The Cancer Genome Atlas Low Grade Glioma [TCGA-LGG] collection. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2016.L4LTD3TK
9. Ronneberger O., Fischer P., Brox T. (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N., Hornegger J., Wells W., Frangi A. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham. https://doi.org/10.1007/978-3-319-24574-4_28
10. Pavel Yakubovskiy. Segmentation models pytorch. https://github.com/qubvel/segmentationmodels.pytorch,2020.
11. Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016. p 2921–2929.
Figures