2128
Mass Univariate Regression Analysis on a Three-Dimensional Liver Phenotype: Associations with Disease States1Research Centre for Optimal Health, School of Life Science, University of Westminster, London, United Kingdom, 2Calico Life Sciences LLC, South San Francisco, CA, United States
Synopsis
Organ MRI measurements have the potential to enhance our understanding of the precise phenotypic changes underlying many clinical conditions. Using liver mesh-based shape analysis we were able to detect significant changes in organ shape in specific anatomical regions associated with anthropometric traits and health conditions including type-2 diabetes and liver disease. We have shown that presence of T2D is associated with increase in liver shape and size. We also showed that the liver disease when accounting for liver fat showed more negative associations with the 3D liver shape phenotype.
Introduction
Magnetic resonance imaging (MRI) has become the benchmark for clinical research in the study of body composition. The incidence of chronic conditions such as type-2 diabetes (T2D), cardiovascular disease and liver disease are rising rapidly, which reflects the increasing prevalence of obesity in society1. Image-derived phenotypes (IDPs) of abdominal organs from MRI, is gaining traction as a means of representing detailed three-dimensional (3D) mesh-derived phenotypes related to variations in shape and may be used to model changes in a normal versus pathological organ and improve diagnosis.Methods
From the whole UK Biobank participants who underwent imaging visits, we selected subjects with type 2 diabetes mellitus (T2D) and those with liver disease (LD) using the codes described previously2. We then identified a control cohort with no disease reported and designed a case-control study for each disease population, where the controls were chosen by matching one individual with every case by a factor of age, gender and body mass index (BMI).We made use of liver segmentations derived from Dixon MRI acquisitions in the UK Biobank, to create 3D liver meshes. Image registration for the construction of the liver template was performed across a gender-balanced population of 200 participants using B-Spline normalisation with mutual information as the similarity metric, and surface meshes were constructed to create the templates in a coordinate space3,4. All subject-specific surfaces were aligned to a template using linear transformations. The template was then warped to each subject’s space using the deformation fields computed via non-rigid transformation using the Image Registration Toolkit implementation5,6.
To determine the differences in the liver surface relative to an average liver shape, such that the outward or inward change would add or subtract from the volume on a region of the liver, a 3D mesh-derived phenotype was created for each subjects mesh. In particular, surface-to-surface (S2S) was measured by computing the signed distance between the template surface and each subject’s surface for each vertex so that positive and negative S2S value indicates an outward or inward change, respectively. A mass univariate regression analysis was then employed to compare the associations between the 3D mesh-derived phenotype and anthropometric variables. We specifically performed a three-dimensional phenotypic regression modeling applied threshold-free cluster enhancement (TFCE) and permutation testing to assess the associations between S2S values from the liver meshes and image-derived phenotypes specific to the liver. This included proton density fat fraction (PDFF) and iron concentration. These were adjusted for relevant covariates with correction to control the false discovery rate, as previously described4. More specifically, in model 1 we used adjustments of age, gender, ethnicity, BMI and waist-to-hip ratio (WHR), in model 2 we included all the covariates from model 1 adding liver PDFF, and in model 3 we included model 2 adding time of the day of the MRI scan.
Results
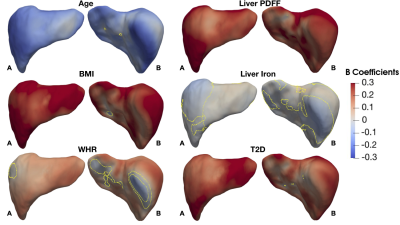
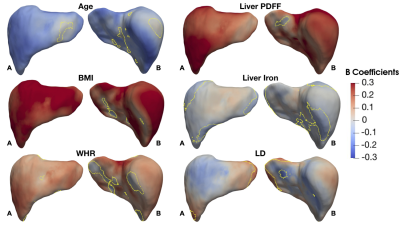
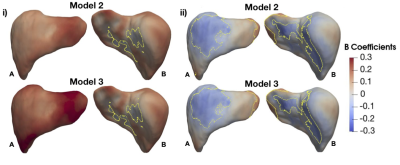
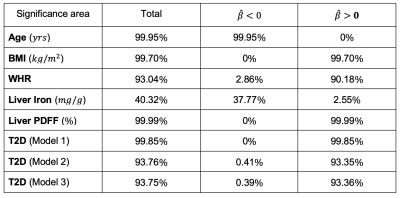
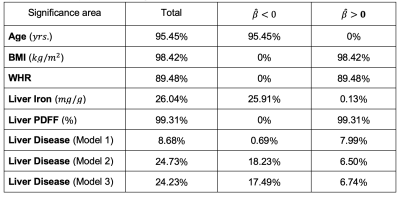
We found 1,911 participants with T2D in UK Biobank from which 67% were male, aged between 46 to 82 years old and a BMI between 18.3 to 50.1 kg/m2 and 479 participants with LD (219F/260M), aged between 48 to 81 years old and a BMI between 18.6 to 43.8 kg/m2. We analysed the liver MRI data to assess the associations between S2S and anthropometric covariates, liver IDPs and disease states for each case-control cohort. As shown in Table 1 and Figure 1, S2S values are shown to decrease with age with significant association on 99.95% of the liver, while WHR, BMI and liver PDFF showed an increase with increased S2S values. Liver iron showed positive associations on 2.55% of the vertices, and negative associations on 37.77%. The presence of T2D was positively associated with S2S on 99.85% of the vertices showing outward shape variations in the liver. However, the significance area was reduced to 93.35 out of 93.75% positive associations when adjusting for liver PDFF. Adding MRI scan time of the day showed minor differences in the significance area (Figure 3i).Similar results were observed between the anthropometric covariates and the liver IDPs in the liver disease cohort. As shown in Figure 2 and Table 2, S2S is shown to decrease with age and increase with BMI, WHR and liver PDFF. Although the presence of liver disease was only positively associated with S2S on 7.99% of the anterior part of the left lobe when adjusting for model 1, we observed negative associations with S2S on 18.23 out of 24.73% and a positive association with liver S2S on 6.50% of the vertices when adjusting for model 2 (Figure 3ii).
Discussion and Conclusion
From the liver surface mesh, we presented S2S values and were able to quantify the anatomical relationships with the anthropometric traits, liver IDPs, and disease states using mass univariate regression analysis. Presence of T2D was associated with outward shape variations in the liver whereas liver disease showed an inward shape change at the anterior part of the right lobe and the posterior parts of the left and right lobes, and an outward increase in liver S2S in the anterior part of the left lobe.Acknowledgements
We thank Matt Sooknah and Amoolya Singh for comments that improved the quality of our work. This research has been conducted using the UK Biobank Resource under Application Number 44584 and was funded by Calico Life Sciences LLC.References
1. Thomas E.L. et al. Metabolic syndrome, overweight and fatty liver. CRC Press 2013; p 763–768
2. Whitcher B. et al. Two years later -- changes in abdominal organs: First observations from the UK Biobank longitudinal imaging study. In press 2021
3. Avants B.B. et al. The optimal template effect in hippocampus studies of diseased populations. Neuroimage 2010; 49: 2457–2466
4. Thanaj, M. et al. Mass Univariate Regression Analysis for Three-Dimensional Liver Image-Derived Phenotypes. MIUA 2021; p 165-176
5. Duan J. et al. Automatic 3D Bi-Ventricular Segmentation of Cardiac Images by a Shape-Refined Multi-Task Deep Learning Approach. IEEE Trans on Med Im 2019; 38: 2151-2164
6. IRTK https://biomedia.doc.ic.ac.uk/software/irtk/
Figures