2090
A Software-based TIAMO approach to enable high resolution large FOV body imaging at 7T ultra-high field1Medical Imaging, Radboud UMC, Nijmegen, Netherlands, 2Erwin L. Hahn Institute for MRI, University Duisburg-Essen, Essen, Germany, 3High-Field and Hybrid MR Imaging, University Hospital Essen, Essen, Germany
Synopsis
Inhomogeneities across large FOVs are problematic at ultra-high field. We propose a software-based TIAMO implementation to enable similar flip angles across the body without hardware interference on the 7T MR scanner. Using an interleaved and complementary CP+/CP2+ excitation scheme, we acquired water-selective and lipid-selective 3D GRE data at high resolution demonstrating homogeneous signal distribution across the pelvis and lower abdomen.
Introduction
Ultra-high field MRI offers high SNR and is therefore an excellent candidate for imaging small anatomical structures such as pelvic lymph nodes (LN) inside the body, which in combination with USPIO nanoparticles can aid in N-staging different types of cancer [1]. A major challenge is the decreased RF wavelength at 7T causing signal dropouts. This can in part be compensated by using RF shimming depending on the number of independent RF transmit channels. However, homogeneous RF shimming over very large field of views (FOV) remains challenging. In the past, the time interleaved acquisition of modes (TIAMO) [2] technique has been proposed to overcome transmit inhomogeneities and enabled large FOV-imaging in the body using an 8-channel parallel transmit system at ultra-high field.TIAMO uses two complementary RF shims in an interleaved fashion to improve overall transmit homogeneity with two complementary excitations reaching similar flip angles across the body [3].
The previous major drawback of this technique was its implementation on designated external RF hardware connected to the MR scanner, which would now interfere with warranty and maintenance issues from the MR system vendor on most recent 7T MR scanners.
In this abstract, we propose a solely software based TIAMO approach which enables high resolution and large FOV imaging at ultra-high field overcoming the mentioned inhomogeneities in the same way as the previously used hardware-based TIAMO solution. As a proof-of-concept, we show water-selective and lipid-selective 7T 3D GRE images of the pelvis.
Methods
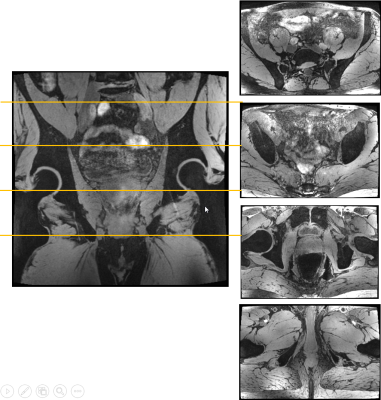
TIAMO was implemented into a standard pTx-capable 3D GRE sequence by modifying the phases of the individual channels of the excitation pulse in the preparation section of the sequence. Two different RF pulses were used. The first pulse was transmitting in CP+ mode for the individual 8 channels, the second pulse used the complementary CP2+ mode. Within the sequence, the two RF pulses were played out in an interleaved fashion using two average settings. Images were reconstructed using 16 “virtual” TIAMO receive elements [3]. Data were acquired from a healthy 47-year old male volunteer after informed consent on a 7T whole body system (Magnetom Terra, Siemens Healthcare GmbH, Germany) running on software version VE12U with a 8Tx/32Rx body-array coil [4].After a localizer and standard B0 shimming, the high resolution protocol could be run without additional adjustments, consisting of two scans:
1. A water excitation scan with a 970 μs block pulse with transmitter frequency centered on the water resonance with the following parameters: 256 coronal partitions, 0.68 mm isotropic resolution, 384x384 matrix, 6/8 partial Fourier (PF) in phase/slice direction, BW 500 Hz/px, PAT 6, TR/TA = 16 ms/9:30 min, 5 gradient echoes with TE(1-5)=2.47/4.79/ 7.11/9.43/11.75 ms.
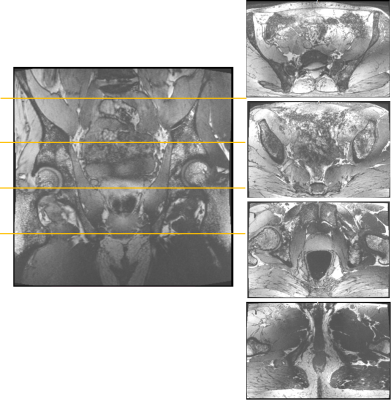
2. A lipid-selective excitation scan with the 970 μs block pulse transmitter frequency centered at the main lipid frequency. Parameters: 256 coronal partitions, 0.68 mm isotropic resolution, 384x384 matrix, 7/8 PF phase/slice, BW 500 Hz/px, PAT 3, TE/TR/TA = 2.48 ms/5.5 ms/7:04 min.
For water-selective excitation, the zero-crossing of the frequency profile of the RF pulse would coincide with the lipid resonance frequency resulting in only water excitation (for the lipid-selective acquisition vice-versa). [5] The signal decay of the multi-gradient echo acquisition was used to calculate any computed echo time image set (example at cTE = 3 ms), an R2* relaxation rate image and an image set with the root-sum-of-squares addition of all signals together.
Results
The 3D water-selective mGRE acquisition provided proper water excitation in the center of the body (Fig. 1). At the central coronal partition, only water is excited as B0 homogeneity is sufficient. Even though the TIAMO shims are not optimized to the volunteer (basic CP+/CP2+ was used), there were no signal dropouts because of missing excitation throughout the large FOV. Without the use of bowel relaxant, intestinal motion causes blur and artefacts. At superficial anterior and posterior parts of the volunteer, lipid excitation is visible caused by B0 inhomogeneity.Observing the same slices for the short TE lipid-selective GRE acquisition it appeared that lipid excitation was more difficult as water had not completely been suppressed (Fig. 2). Contrast between water and lipids is present, although inhomogeneous over the large FOV. Subcutaneous and visceral lipid tissue is very bright and even some lipid signal is visible within the bones.
Discussion and Conclusion
High resolution large FOV images with a reasonably homogeneous signal distribution and high SNR were obtained in the pelvis for lipid- and water-selective acquisitions using a software-based TIAMO implementation. In this work, we used the standard CP+ /CP2+ shim modes, but the flexibility of the software implementation allows to easily incorporate any pair of (universal complementary) shim settings into the sequence. Room for improvement is in better B0-shimming and in estimating the attained flip angle with the two excitations with different B1 shim settings [6]. The concept can also be transferred to other sequences for general use. Furthermore, the software-based approach enables the possibility to easily and independently distribute the TIAMO technique to different sites and MR scanners which was not possible with the previous hardware solution.Acknowledgements
This project has received funding from the European Union in the EFRO
program, project number PROJ-01009. The 7T MRI system at the ELH Institute has
been funded by the Deutsche Forschungsgemeinschaft (DFG, German Research
Foundation) – Project number 432657511.
References
[1] Wu, Liucheng, et al. "Diagnostic performance of USPIO-enhanced MRI for lymph-node metastases in different body regions: a meta-analysis." European journal of radiology 80.2 (2011): 582-589.
[2] Orzada, Stephan, et al. “RF excitation using time interleaved acquisition of modes (TIAMO) to address B1 inhomogeneity in high-field MRI.” Magnetic resonance in medicine 64,2 (2010): 327-33.
[3] Orzada, Stephan, et al. "Mitigation of B1+ inhomogeneity on single‐channel transmit systems with TIAMO." Magnetic resonance in medicine 70.1 (2013): 290-294.
[4] Rietsch, Stefan HG, et al. "7T ultra‐high field body MR imaging with an 8‐channel transmit/32‐channel receive radiofrequency coil array." Medical physics 45.7 (2018): 2978-2990.
[5] Philips, Bart WJ, et al. "High resolution MR imaging of pelvic lymph nodes at 7 Tesla." Magnetic resonance in medicine 78.3 (2017): 1020-1028.
[6] Brunheim, Sacha, et al. “Fast and accurate multi-channel B1+ mapping based on the TIAMO technique for 7T UHF body MRI.” Magnetic resonance in medicine 79.5 (2018): 2652-2664.
Figures