2048
Mild neuroanatomical phenotype in an ALS/FTD mouse model with mutation in C9orf721Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 2UK Dementia Research Institute at UCL, Faculty of Brain Sciences, University College London, London, United Kingdom, 3Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom, 4Mouse Imaging Centre, The Hospital for Sick Children, Toronto, ON, Canada, 5Mammalian Genetics Unit, MRC Harwell Institute, Oxfordshire, United Kingdom, 6Department of Neuromuscular Diseases, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom
Synopsis
Mutations in the C9orf72 gene are the most prevalent genetic alteration in ALS/FTD. This study investigates neuroanatomical phenotypes in two C9orf72 knock-in mouse models separately expressing either poly-(PR) or poly-(GR) dipeptide-repeats. Ex-vivo structural MRI (40 μm isotropic resolution) was acquired at 7T. After registration, the deformation fields were used to estimate voxels and region-of-interest volumes for comparison between mutants and wild-type mice. Although neuroanatomical phenotypes have been previously described in C9orf72 patients and other ALS/FTD mouse models, 20-month-old poly-(PR) and poly-(GR) mice presented subtle alterations. Further investigations to assess microstructural and histological changes in these mouse models are in progress.
Introduction
Mutations in the C9orf72 gene are the most prevalent genetic alteration in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) patients1,2. Specifically, patients present a repeat expansion of a six-nucleotide sequence in the C9orf72 gene, ranging from 30 to several thousand repeats3. Although this sequence is in a non-coding region of the gene, it can be unconventionally translated into dipeptide repeat proteins (DPRs). DPR inclusions are observed in the brain of FTD and ALS patients early in the disease4. From all five possible DPRs translated from this repeat expansion in C9orf72, at least two, poly-(GR) and poly-(PR), are known to be potently neurotoxic5.In this study, ex-vivo MRI was used to assess possible neuroanatomical alterations in two C9orf72 knock-in mouse models separately expressing either poly-(PR) or poly-(GR) DPRs. These mice were recently generated using CRISPR technology to target the endogenous mouse C9orf72 locus and insert codon-optimised repeats in frame with the endogenous ATG, leading to expression of either poly-(PR) or poly-(GR) peptides under the control of the endogenous mouse C9orf72 promoter. Our hypothesis was that these mice would develop FTD- and/or ALS-like neuroanatomical phenotypes that could be captured with ex-vivo MRI. Here, we present first results from structural MRI, with microstructural MRI and histology planned as future work on the same cohorts.
Materials and Methods
Ten poly-(PR), 8 poly-(GR) mice and 10 wild-type (WT) littermates (females, 20.4±0.8 months-old) were studied. Mice were deeply anesthetized with isoflurane and intracardially perfused with saline solution, followed by 4% formalin. Both saline and formalin contained 2 mM Gd-contrast agent (Gd-CA; Gadovist, Bayer Vital GmbH, Leverkusen, Germany). Heads were removed and dissected, keeping brains in the skull. Brains were immersed in 4% formalin with 2 mM Gd-CA for 24h at 4oC, and then kept in PBS with 2 mM Gd-CA and 0.05% azide at 4oC until scanned. MRI was performed on a 7.0 tesla MRI scanner (Agilent Inc., Palo Alto, CA), using sixteen custom-built solenoid coils to image the brains in parallel6. Anatomical images with 40 μm isotropic resolution were acquired using a T2-weighted 3D fast spin-echo sequence, with TR=350 ms, 6 evenly spaced echoes between TE=12-72 ms (effective TE=30ms), field-of-view of 20x20x25 mm3, matrix size = 504x504x630, 4 averages and cylindrical acquisition of k-space7. Total imaging time was approximately 14 hours.Brain images were iteratively aligned to a minimum deformation template space generated from all the brains in the study8,9, and the deformation field necessary to bring each brain to this space was used to compute voxel volumes, represented as the logarithm of the Jacobian determinant. Voxel volumes were used to compute the total brain volume for each mouse and the volume of 182 regions of interest after atlas-based segmentation10. Volumes were modelled as a function of genotype, and false discovery rate (FDR) was used for multiple comparison correction.
Results
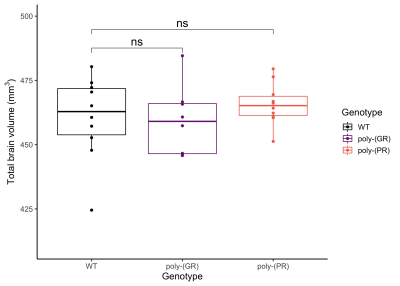
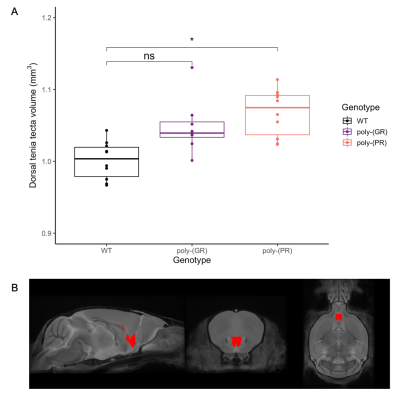
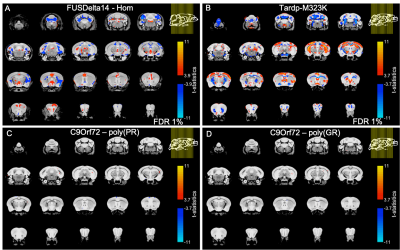
There was no detectable effect of genotype in total brain volume (Figure 1). On the analysis of regions of interest after atlas-based segmentation, only one brain region, the dorsal taenia tecta, presented a significant difference in volume as a function of genotype. The volume of the dorsal taenia tecta was increased in poly-(PR) mice when compared to WT. Poly-(GR) mice presented a trend of increased volume in the same region, but it didn’t reach statistical significance (Figure 2). No significant differences could be identified on the voxel-wise analysis for poly-(PR) or poly-(GR) mice (FDR 20%, Figure 3).Discussion
Cortical thinning11 and white matter alterations12 are generally described in ALS/FTD, and widespread cortical and subcortical atrophy is observed in C9orf72 patients and pre-symptomatic carriers13. Interestingly, there are also reports of increased grey matter volume in ALS patients without cognitive impairment12, which has been related to inflammation preceding neuronal loss or to compensatory mechanisms.Our group has previously reported widespread relative volumetric changes in the brain of two mouse models of ALS/FTD with mutation in the Fus gene14 (FUSDelta14-homozygous15) and in the Tardp gene16 (Tardp-M323K17). In contrast to the C9orf72 models described here, those models exhibited multiple regions of decreased and increased relative volume when compared to WT littermates (Figure 3, A-B).
Mouse models with mutation in C9orf72 are relevant for ALS/FTD since they correspond to the most common genetic cause of these diseases1,2. However, in the present study, there were only mild neuroanatomical alterations in the poly-(PR) mouse model with mutation in C9orf72, and no detectable change in poly-(GR) mice. The increased volume in the dorsal taenia tecta, a grey matter region in the olfactory peduncle, could reflect inflammation and oedema in this area due to poly-(PR) DPR toxicity. Other MRI modalities exploring tissue microstructure, as well as histological analysis will allow further exploration of the brain phenotypes in these C9orf72 DPR knock-in mouse models.
Conclusions
Mouse models with mutation in C9orf72 leading to selective expression of poly-(GR) and poly-(PR) DPRs presented a subtle neuroanatomical phenotype, with volumetric alterations detected with MRI only in poly-(PR) mice. Further investigations to assess microstructural alterations in these mouse models of ALS/FTD are in progress. Histological analysis will allow to identify the microscopic alterations leading to MRI phenotypes.Acknowledgements
Brian J. Nieman, Jason Lerch and Karla Miller contributed equally to this work. This work was supported by the Wellcome Trust (grant 202788/Z/16/Z), Oxford Wellcome Institutional Strategic Support Fund (0009875), The Motor Neurone Disease Association and the Packard Center for ALS Research. The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (grant 203139/Z/16/Z).References
1. Renton, A. E. et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 72, 257–268 (2011).
2. DeJesus-Hernandez, M. et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 72, 245–256 (2011).
3. van Es, M. A. et al. Amyotrophic lateral sclerosis. Lancet 390, 2084–2098 (2017).
4. Freibaum, B. D. & Taylor, J. P. The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci. 10, 1–9 (2017).
5. Balendra, R. & Isaacs, A. M. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 14, 544–558 (2018).
6. Bock, N. A., Nieman, B. J., Bishop, J. B. & Henkelman, R. M. In vivo multiple-mouse MRI at 7 Tesla. Magn. Reson. Med. 54, 1311–1316 (2005).
7. Spencer Noakes, T. L., Henkelman, R. M. & Nieman, B. J. Partitioning k -space for cylindrical three-dimensional rapid acquisition with relaxation enhancement imaging in the mouse brain. NMR Biomed. 30, e3802 (2017).
8. Nieman, B. J. et al. MRI to Assess Neurological Function. Curr. Protoc. Mouse Biol. 8, e44 (2018).
9. Friedel, M., van Eede, M. C., Pipitone, J., Chakravarty, M. M. & Lerch, J. P. Pydpiper: a flexible toolkit for constructing novel registration pipelines. Front. Neuroinform. 8, 1–21 (2014).
10. Chakravarty, M. M. et al. Performing label-fusion-based segmentation using multiple automatically generated templates. Hum. Brain Mapp. 34, 2635–2654 (2013).
11. Verstraete, E. et al. Structural MRI reveals cortical thinning in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 83, 383–388 (2012).
12. Christidi, F. et al. Gray matter and white matter changes in non-demented amyotrophic lateral sclerosis patients with or without cognitive impairment: A combined voxel-based morphometry and tract-based spatial statistics whole-brain analysis. Brain Imaging Behav. 12, 547–563 (2018).
13. Li Hi Shing, S. et al. The imaging signature of C9orf72 hexanucleotide repeat expansions: implications for clinical trials and therapy development. Brain Imaging Behav. 15, 2693–2719 (2021).
14. Bach, A. B. M. et al. Brain structure in the homozygous FUSDelta14 mouse recapitulates amyotrophic lateral sclerosis phenotypes. in Proc. Intl. Soc. Mag. Reson. Med. 28 0899 (2020).
15. Devoy, A. et al. Humanized mutant FUS drives progressive motor neuron degeneration without aggregation in ‘FUSDelta14’ knockin mice. Brain 140, 2797–2805 (2017).
16. Martins-bach, A. B. et al. Anatomical and microstructural brain alterations in the TDP-M323K mouse model of amyotrophic lateral sclerosis. in Proc. Intl. Soc. Mag. Reson. Med. 29 1208 (2021).
17. Fratta, P. et al. Mice with endogenous TDP‐43 mutations exhibit gain of splicing function and characteristics of amyotrophic lateral sclerosis. EMBO J. 37, e98684 (2018).
Figures