1980
Erwin - a quantitative MRI toolbox1ICube, Université de Strasbourg-CNRS, Strasbourg, France
Synopsis
Erwin is an open-source Python toolbox dedicated to the computation of quantitative maps from MRI data. It provides a unified interface, through either its Python API or its command-line tool, to well-known qMRI methods (e.g. B1 mapping using Actual Flip Angle (AFI) or T1 mapping using Variable Flip Angle (VFA)) and external toolboxes (e.g. MRtrix or MEDI). Erwin also provides tools to create complex pipelines where the dependencies between steps are tracked: a change in a source image or a method parameter will only trigger the recomputation of a minimal set of subsequent steps.
Introduction
Quantitative MRI, i.e. measuring biophysical parameters using MRI, is a very active research field and can be traced back to the early days of NMR with Hahn's method to measure T21. In this abstract, we describe a new Python-based toolbox, also providing command-line tools, a dependency handler to easily define complex pipelines, and access to imaging meta-data, including vendor-specific fields.Design and features
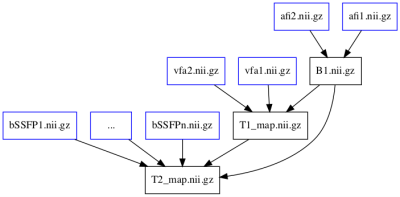
To balance the ease of use with the potentially complex qMRI pipelines, Erwin has two main interfaces: a set of command line tools and a Python API. For quick tests or for the simpler methods, the command-line tools hide the details of the Python API in a unified syntax, reachable by the main executable "erwin": a list of all methods can be obtained by calling "erwin --list", the details of each method and the corresponding arguments are given by calling e.g. "erwin b1_map.afi --help". These command-line tools are automatically defined from the Python API, which ensures feature parity between the two interfaces. The Python API will prove useful for more complex pipelines, e.g. the T2 mapping based on bSSFP2 which requires, in addition to a set of bSSFP images with varying flip angles and phase increments, a B1-map and a T1-map. This is accomplished in Erwin by defining the inputs and outputs of each method, and by connecting the output of a task to the input of the next one, as shown in Figure 1; in this example, the T1-map is computed using the VFA method3, which requires the B1 map. This effectively builds a directed dependency graph between steps, with the property that changing one of the steps does not require recomputing the upstream steps, only the downstream ones: in the previous example, replacing the T1-map will only recompute the T2-map, while replacing the B1-map, also used in the T1-map, will recompute first the T1-map, then the T2-map, ensuring the result is always up to date, with a minimal computation cost. The pipeline engine in Erwin also allows to generate a pictorial representation of a task graph, shown in Figure 2 for the example of the T2-map pipeline.One common characteristic of many qMRI methods is their use of acquisition parameters, e.g. flip angle, repetition time or b-value. These parameters are usually stored either in the DICOM files produced by the scanner or in separate meta-data files such as the BIDS sidecar files. In the case of DICOM, some of the parameters are included only in vendor-dependent fields, as is the case for the phase increment of the T2-bSSFP method cited above. Moreover, not all formats use the same unit for a given field: DICOM store the timing parameters (echo time, repetition time, inversion time, etc.) in milliseconds, while BIDS uses seconds. In order to be vendor-independent, Erwin provides tools to access meta-data stored in either DICOM or JSON format, with the option to convert it from its specific unit. For example, the inversion times of a pulsed ASL sequence4, stored in the Siemens private field using microseconds can be accessed by "erwin meta_data.get -p XXX.dcm -q 00291020.MrPhoenixProtocol.0.alTI".
Erwin does not strive to re-implement all qMRI methods: when a complete and well-known toolbox exists for a specific set of methods, e.g. MRtrix5 for diffusion or MEDI6 for QSM, the provided tools are simply wrapped as tasks so that they can transparently be included in pipelines. This is straightforward for pre-compiled executables or Python libraries, and, for MATLAB-based code, we provide a bridge between the MATLAB engine and the Python runtime.
Conclusion
Erwin has been released under free software license (MIT), is available on GitHub7 and PyPi8, and includes a documentation of all provided methods9, and a set of reference images with their meta-data10. The methods included as of the writing of this abstract target field mapping (B0 and B1), relaxometry (T1, T2 and T2*), cerebral blood flow, diffusion, magnetization transfer, and susceptibility. Although obviously not complete due to the ever-growing number of publications regarding qMRI, the features already present in Erwin and its open development model can benefit the whole qMRI community.Acknowledgements
No acknowledgement found.References
1.
Hahn. Spin Echoes. Physical Review 80(4). 1950
2. Jutras et al. Analytical corrections of banding artifacts in driven
equilibrium single pulse observation of T2. MRM 76(6). 2016
3. Preibisch & Deichman. Analytical corrections of banding artifacts in
driven equilibrium single pulse observation of T2. MRM 62(1). 2009
4. Wang et al. Pediatric perfusion imaging using pulsed arterial spin labeling.
JMRI 18(4). 2003
5. Tournier et al. MRtrix3: A fast, flexible and open software framework for
medical image processing and visualisation. NeuroImage, 202. 2019
6. http://pre.weill.cornell.edu/mri/pages/qsm.html
7. https://github.com/lamyj/erwin
8. https://pypi.org/project/erwin
9. https://erwin.readthedocs.io
10. https://iris.icube.unistra.fr/nextcloud/s/iYX7pgHNeAp7FF6/download