1967
Longitudinal Multiple Sclerosis Lesion Segmentation Using Pre-activation U-Net1Department of Electrical Engineering (ESAT), KU Leuven, Leuven, Belgium, 2CREATIS (CNRS UMR5220 & INSERM U1294), Université Claude-Bernard Lyon 1, Lyon, France
Synopsis
Automated segmentation of new multiple sclerosis (MS) lesions in MRI data is crucial for monitoring and quantifying MS progression. Manual delineation of such lesions is laborious and time-consuming since experts need to deal with 3D images and numerous small lesions. We propose a 3D encoder-decoder architecture with pre-activation blocks to segment new MS lesions in longitudinal FLAIR images. We also applied intensive data augmentation and deep supervision to mitigate the limited data and the class imbalance problem. The proposed model, called Pre-U-Net, achieved a Dice score of 0.62 and a sensitivity of 0.58 on the public challenge MSSEG-2 dataset.
Introduction
Multiple sclerosis (MS) is a common chronic, autoimmune demyelinating disease of the central nervous system, which causes inflammatory lesions particularly in white matter (WM). Multi-parametric MRI constitutes the main imaging tool in clinical practice, to diagnose and assess MS lesion load, using mainly, FLuid Attenuated Inversion Recovery (FLAIR) images to distinguish WM lesions appearing as high-intensity regions. It is highly relevant to monitor lesion activity, and especially the appearance of new lesions or the enlargement of existing lesions, for various purposes, including prognosis and follow-up. Lesional changes between two longitudinal MRI scans from an MS patient are the most important markers for tracking disease progression and inflammatory changes. Thus, the accurate segmentation of new lesions is an essential prerequisite for quantifying MS progression and measuring features, such as lesion volume and location. However, manual annotation of such lesions is tedious and time-consuming especially because raters need to deal with multiple 3D images for each case. Furthermore, longitudinal MS lesion segmentation is very challenging due to the very small size of new lesions and their large inter-patient variability in shape, size, and location. Therefore, accurate computer-assisted tools are needed to automatically perform such tedious clinical tasks.Methods
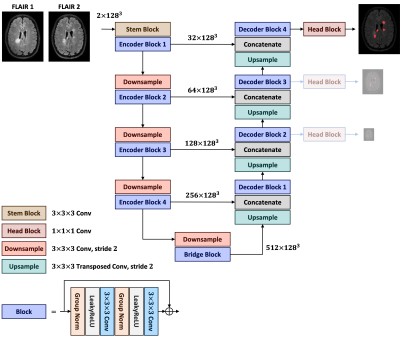
Architecture. The proposed model, called Pre-U-Net, follows a U-Net-style1 architecture made up of encoder and decoder parts (see Figure 1). A 3x3x3 convolution is used as the stem layer. The network takes a 2-channel image of size 128x128x128 and outputs a probability map with the same spatial size. The network encoder (decoder) has 4 levels, at each of which the input is downsampled (upsampled) by a factor of two while the depth is doubled (halved). Downsampling and upsampling are performed via strided convolution and strided transposed convolution, respectively. We used deep supervision2 at the three highest resolutions in the decoder, applying pointwise convolutions to get three auxiliary logit tensors. The cornerstone of our model is Pre-activation ResNet block3, which is composed of two 3x3x3 convolutions, each of which follows LeakyReLU activation and Group Normalization (with a group size of 8). A pointwise convolution may be used in the shortcut connection to match the input dimension with the output dimension of the residual mapping. This Pre-U-Net has 26.3 million trainable parameters.Preprocessing. For each patient, we first concatenated the two FLAIR images to form a 2-channel 3D image as the input. Each image and its ground truth were then cropped with a minimal box filtering out zero regions. We normalized each image channel-wise using z-score to have intensities with zero mean and unit variance. All the images and their ground truths were then resampled to the same voxel spacing of 0.6 mm3 using trilinear interpolation. Finally, we cropped random 128x128x128 patches and performed oversampling from lesion regions such that 50% of the patches contained some lesion to cope with the class imbalance problem.
Data Augmentation. To reduce overfitting, we passed the data through a data augmentation workflow before feeding it into the network. We applied random spatial transforms, including affine and flip, and random intensity transforms, including Gaussian smoothing and intensity shifting.
Optimization. All networks were trained for 100k steps with batch size 2 using AdamW optimizer with the initial learning rate of 10-5, weight decay of 10-2, and a cosine annealing scheduler. The loss function was the sum of soft Dice and Focal loss. The three deep supervision outputs and the corresponding downsampled ground truths were used for loss computation. The training objective function was the weighted mean of the losses at all resolutions, with the weights being 1, 0.5, and 0.25 at resolutions 1283, 643, and 323, respectively.
Inference. A test image in the inference was first subjected to z-score intensity normalization and resampled to a voxel spacing of 0.6 mm3. The prediction was then made using a sliding window approach with a 50% overlap and a window size of 128x128x128. We resampled the resulting logit tensor back to the original voxel spacing and finally thresholded it to obtain a binary segmentation map.
Experiments
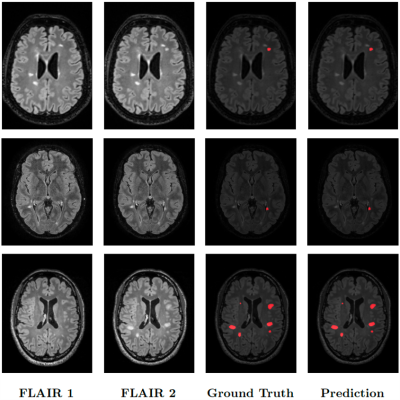
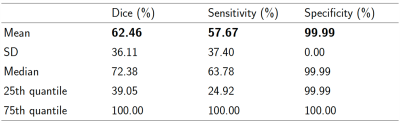
Data. The MSSEG-24,5 public dataset consisted of forty 3D FLAIR images acquired at two time points and co-registered in the intermediate space between the two time points. New lesions were manually annotated by multiple raters, and the consensus ground truths were obtained through a voxel-wise majority voting (see Figure 2).Results. We performed 5-fold cross-validation to verify the model's capability to generalize to unseen data. The scores are reported in Table 1. Particularly, Pre-U-Net could achieve a Dice score of 0.62 and a sensitivity of 0.58. Figure 2 shows some qualitative results.
Discussion and Conclusion
The smaller value of sensitivity and higher value of specificity indicate a tendency to undersegmentation, which is attributed to the problem of class imbalance and data insufficiency. The obtained scores may appear too small but were actually ones of the best of the challenge, showing its level of difficulty.In conclusion, this work proposed a U-Net-style architecture consisting of pre-activation blocks, called Pre-U-Net, which performed very effectively in the difficult constraints imposed by the challenge. This offers a promising automatic tool for new MS lesion segmentation, which is crucial for monitoring disease progression and helping neurologists in clinical practice.
Acknowledgements
The research leading to these results has received funding from EU H2020 MSCA-ITN-2018: INtegrating Magnetic Resonance SPectroscopy and Multimodal Imaging for Research and Education in MEDicine (INSPiRE-MED), funded by the European Commission under Grant Agreement #813120. This research also received funding from the Flemish Government (AI Research Program). Sabine Van Huffel and Pooya Ashtari are affiliated to Leuven.AI - KU Leuven institute for AI, B-3000, Leuven, Belgium.References
1. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention 2015 Oct 5 (pp. 234-241). Springer, Cham.
2. Lee CY, Xie S, Gallagher P, et al. Deeply-supervised nets. Artificial intelligence and statistics 2015 Feb 21 (pp. 562-570). PMLR.
3. He K, Zhang X, Ren S, et al. Identity mappings in deep residual networks. European conference on computer vision 2016 Oct 8 (pp. 630-645). Springer, Cham.
4. Vukusic S, Casey R, Rollot F, et al. Observatoire Français de la Sclérose en Plaques (OFSEP): A unique multimodal nationwide MS registry in France. Multiple Sclerosis Journal. 2020 Jan;26(1):118-22.
5. Confavreux C, Compston DA, Hommes OR, et al. EDMUS, a European database for multiple sclerosis. Journal of Neurology, Neurosurgery & Psychiatry. 1992 Aug 1;55(8):671-6.
Figures