1964
MR-Class: MR Image Classification using one-vs-all Deep Convolutional Neural Network1CCU Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Medical Faculty, Heidelberg University Hospital, Heidelberg, Germany, 3Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg, Germany, 4German Cancer Consortium (DKTK) Core Center, Heidelberg, Germany, 5Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany, 6CCU Radiation Therapy, German Cancer Research Center (DKFZ), Heidelberg, Germany, 7National Center for Tumor Diseases (NCT), Heidelberg, Germany
Synopsis
MR-Class is a deep learning-based MR image classification tool that facilitates and speeds up the initialization of big data MR-based research studies by providing fast, robust, and quality-assured MR image classifications. It was observed in this study that corrupt and misleading DICOM metadata could lead to a misclassification of about 10%. Therefore, in a field where independent datasets are frequently needed for study validations, MR-Class can eliminate the cumbrousness of data cohorts curation and sorting. This can greatly impact researchers interested in big data multiparametric MRI studies and thus contribute to the faster deployment of clinical artificial intelligence applications.
Introduction
Radiomics 1 analyses or artificial intelligence (AI) applications on brain images often require specific magnetic resonance (MR) images, e.g., for the automatic segmentation of glioblastomas. However, in large retrospective datasets collected from multiple scanners and hospitals, medical image classification is complicated by inconsistent naming schemes and missing DICOM metadata 2. Therefore, the automatization and standardization of the retrieval and classification of MR images would be beneficial in terms of time efficiency and accuracy. Deep convolutional neural networks (CNN) in medical image analysis have achieved state-of-the-art performance3, notably when dealing with MR image classification4-5. However, a limitation of the available methods is the inability to deal with the open-set recognition 6, a common issue when dealing with retrospective cohort studies. Thus, we aimed to establish a CNN-based classification method, MR-Class, for the efficient automatic classification of MR images, with the capability of handling unknown classes.Methods
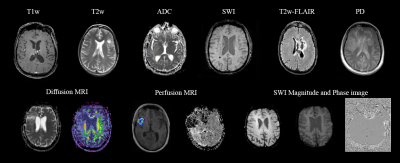
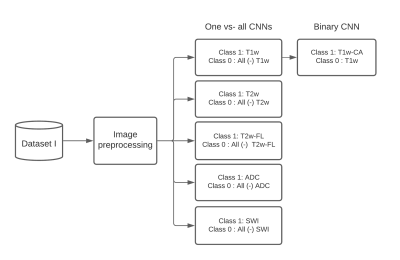
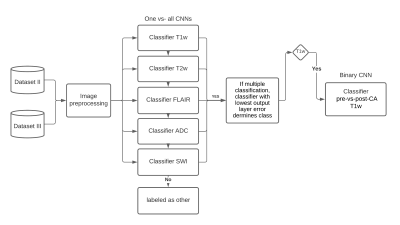
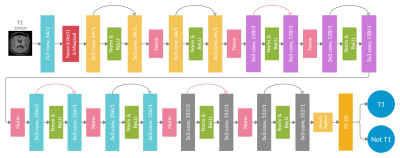
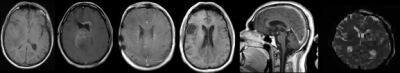
A dataset of 20101 MR images, with 28 different MR scan types, from 320 patients with high-grade glioma (HGG) was used for training. After adequate image preprocessing, all images found in the dataset were included in the training. Still, the classifiable MR images are pre-and post-contrast agent (CA) T1w, T2w, T2w fluid-attenuated inversion recovery (FLAIR), apparent diffusion coefficient (ADC), and susceptibility-weighted imaging (SWI). Sample images found in the cohorts can be seen in Fig.1. MR-Class consists of 5 stacked one-vs-all ResNet7 CNNs, each trained to identify a single MR scan type, followed by a binary classifier for T1w images to determine whether contrast agent (CA) is present. A summary of the training workflow can be seen in Fig.2. Testing was on 11333 images (dataset II), with 22 different MR scan types, from 197 recurrent HGG patients and 3522 images (dataset III), with 17 different MR scan types, from 256 glioblastomas (GBM) patients from the Cancer Genome Atlas (TCGA-GBM) data collection. The MR images are fed to each CNN classifier to infer the corresponding class. If an image is labelled by more than one classifier, the classifier with the lowest output layer error determines the class, while if none of the classifiers labels an image, it is rendered unclassifiable. A summary of the inference workflow is shown in Fig.3. The one-vs-all classification approach is implemented in an attempt to deal with the open-set recognition problem and thus to enable the handling of unknown classes (A schematic representation is shown in Fig.4).Results
Overall accuracy on the independent datasets was 96.7% [95% CI: 95.8, 97.3] for dataset II and 94.4% [95% CI: 93.6, 96.1] for dataset III, i.e., out of 15266 MR images, 620 were misclassified. Furthermore, 96.1% (dataset II) and 92.2% (dataset III) of the MR images not belonging to one of the six classifiable classes were correctly labelled as unknown. Most of the falsely positive labelled images were misclassified diffusion-weighted imaging (DWI) scans as T2w and T1w-FLAIR images labelled as T2w-FLAIR. All other misclassifications can be sorted into different categories: the presence of high anatomical abnormalities, MR artefact-blurring, MR artifacts-other, similar image content for different MR images, and DICOM corrupted scans. Examples of misclassified images are shown in Fig.5.Discussion
This work presents an MR image classifier, MR-Class, which identifies between T1w pre- and post-Ca, T2w, FLAIR, ADC, and SWI while handling unknown classes with an average accuracy of 95.5% on two independent datasets. MR-Class design enables the handling of unknown classes since each CNN only classifies an image if it belongs to its respective scan type, and thus an image not labelled by any of the CNN is rendered as unknown. MR-Class is a helpful tool for data preparation since it eliminates the need to manually sort out the images, which is tedious due to a large amount of data and the use of different naming schemes. Furthermore, since MR-Class classifies the images based on the content rather than the metadata, any corrupted image would be automatically disregarded, i.e. all images labelled as a specific class would have similar content, which is highly beneficial for AI applications.Conclusion
To summarize, we provide a CNN-based classifier for the automatic classification of MR images that could facilitate the initialization of MR-based research studies and AI application development.Acknowledgements
Funding by the H2020 MSCA-ITN PREDICT project.References
1. Lambin, P., Rios-Velazquez, E., Leijenaar, R., Carvalho, S., van Stiphout, R.G.P.M., Granton, P., Zegers, C.M.L., Gillies, R., Boellard, R., Dekker, A., Aerts, H.J.W.L.: Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 48, 441–446 (2012). https://doi.org/10.1016/J.EJCA.2011.11.036
2. Gueld, M.O., Kohnen, M., Keysers, D., Schubert, H., Wein, B.B., Bredno, J., Lehmann, T.M.: Quality of DICOM header information for image categorization. In: Medical imaging 2002: PACS and integrated medical information systems: design and evaluation. pp. 280–287 (2002)
3. Shen, D., Wu, G., Suk, H.-I.: Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 19, 221–248 (2017)
4. Remedios, S., Pham, D.L., Butman, J.A., Roy, S.: Classifying magnetic resonance image modalities with convolutional neural networks. In: Medical Imaging 2018: Computer-Aided Diagnosis. p. 105752I (2018)
5. van der Voort, S.R., Smits, M., Klein, S.: DeepDicomSort: An Automatic Sorting Algorithm for Brain Magnetic Resonance Imaging Data. Neuroinformatics. 19, 159–184 (2021)
6. Scheirer, W.J., de Rezende Rocha, A., Sapkota, A., Boult, T.E.: Toward open set recognition. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1757–1772 (2012)
7. He, K., Zhang, X., Ren, S., Sun, J.: Deep Residual Learning for Image Recognition. (2015)
Figures