1960
Registration of abdominal DCE-MRI: optimal choice of reference and moving image pairs1University of Edinburgh, Edinburgh, United Kingdom, 2Canon Medical Research Europe, Edinburgh, United Kingdom
Synopsis
Before registration can take place, an approach to pairing images must be chosen. We used synthetic data to provide a ground truth to evaluate four approaches to pairing images. Our synthetic data was created using tracer kinetics modelling of abdominal DCE-MRI to create motionless data. Then local distortions were applied to the motionless data. We found that using one image as a reference image produced the best result to eliminate the propagation of errors. The reference image must have a similar intensity distribution to the other images in the sequence.
Introduction
The registration of dynamic contrast enhanced – MRI (DCE-MRI) is challenging due to the complexity of the data. The change in intensity in the voxels can be mistakenly identified as motion, resulting in poor registration. When comparisons are made between pre and post contrast images, new features can be present in the post contrast images causing registration algorithms to fail1. An important stage in the registration process is choosing the pair of images to be registered. Most common approaches will select one image in the series to act as the reference image for all the others. However, there are disadvantages for the selection of any image, such as, difference of intensities between images or the presence of features from the contrast agent (CA). An alternative approach is to register pairs of images adjacent in time through the series, but any errors created during the registration process can be propagated, lowering the accuracy of the registration. Moreover, the lack of ground truth (GT) seriously hinders the estimation of this accuracy. In this work, a new validation scheme is proposed which paired similar images together, but reduced the chance of propagating errors throughout the series. A new validation scheme based on synthetic data is proposed to provide a GT to evaluate our approach with other common methods of pairing images.Methods
We created 56 2D motionless DCE-MR datasets using tracer kinetics (TK) modelling to fit a two compartmental uptake model2 to each voxel in the unregistered data following the method of Reavey et al.3. We chose the central slice from 3D data which usually contains the uterus at its largest point in the image. From this we obtained values for the extraction fraction, plasma flow and the plasma volume, which were used to create motion-free time intensity curves. A synthetic deformation field using gaussian and sinusoidal displacements was created. These distortions were placed randomly in the image ensuring no overlapping occurred. This was achieved by ensuring that the distance of the warp did not exceed the distance between placement of distributions. Deformations were applied to the image, enabling registration, and comparison of accuracy between approaches. Based on previous experiments, SyN4 from the ANTs package was used for registration. The mean squared error (MSE) between the registered imaged and the GT image was calculated to assess registration accuracy. Multiple schemes of images pairing were tested. First, the pre-contrast image was used as the reference image. The second experiment saw the last post-contrast image defined as the reference image while the final experiment paired adjacent images in the time series. Using information learned from the above tested pairings, an approach aiming to pair similar images and reduce the possibility of compounding errors was used. Five images were selected in the time series to be reference images for several subsequent images in the series. The selected reference images were registered together with the remaining images registered to the previously registered images.Results
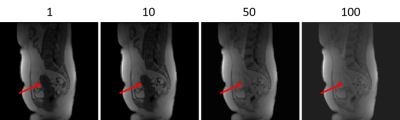
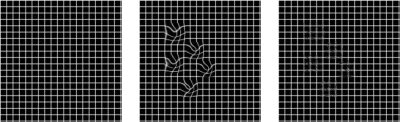
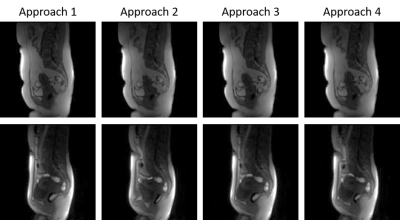
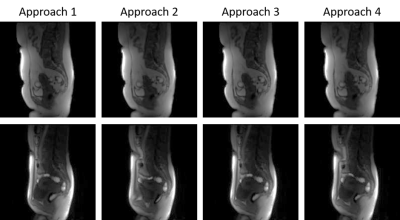
Figure 1 shows an example of multiple timepoints from motionless data created from TK modelling. The uterus has an increase in intensity. Figure 2 shows a displacement field applied to an image.Figure 3 shows results of using different approaches to registration. The MSE with its standard deviation is shown for all images and for images placed into groups of 15 based on their location in the series. Figures 3 and 4 shows example results from each approach in two patients. Figure 4 shows the result when there is no CA present and figure 5 shows the result with CA present.
Discussion
The results of this experiment provided quantitative evaluation of multiple approaches to image pairing when registering DCE-MRI. The first approach performed well when registering the first 15 images in the series due to there being no CA present. However, when there was CA present, the accuracy fell. Overall, this approach scored a MSE of 14.05 ± 5.11. The second approach followed the same trend as the previous approach but in the opposite direction. The first 15 images were registered poorly, however, when CA was present, the accuracy of the registration increased. This enabled for a better overall accuracy than the first approach giving a MSE of 5.37 ± 4.46. The third approach showed that when registering adjacent images, errors can be propagated throughout giving lower accuracy. Due to this, as the registration proceeded throughout the series, the accuracy was lowered at each step giving an overall MSE of 60.39 ± 31.99. The final approach showed an increase in performance for image ranges 121-150. The other images saw a decrease in accuracy due to the propagation of errors throughout the registration process.Conclusion
The results clearly show that if CA is present, using an image with CA present as reference is the best choice. For images without contrast (or with negligible contrast) the non-contrast image is better. Since most of the images in the data have CA present, the contrast image is a better choice. Using adjacent images is poor because as expected the error accumulates throughout the sequence. When registering time series data, selecting one reference image gives optimal results.Acknowledgements
This work was funded through a PhD scholarship by Medical Research Scotland and Canon Medical Research Europe.References
1. Buonaccorsi, G. A. et al. Tracer kinetic model-driven registration for dynamic contrast-enhanced MRI time-series data. Magn. Reson. Med. 58, 1010–1019 (2007).
2. Brix, G. et al. Microcirculation and microvasculature in breast tumors: Pharmacokinetic analysis of dynamic MR image series. Magn. Reson. Med. 52, 420–429 (2004).
3. Reavey, J. J. et al. Markers of human endometrial hypoxia can be detected in vivo and ex vivo during physiological menstruation. Hum. Reprod. (2021) doi:10.1093/humrep/deaa379.
4. Avants, B. B., Epstein, C. L., Grossman, M. & Gee, J. C. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008).
Figures