1952
Think outside the box: Exploiting the imaging workflow for Deep Learning based motion estimation and correction1Pattern Recognition Lab Friedrich-Alexander-University Erlangen-Nuremberg, Erlangen, Germany, 2Erlangen Graduate School in Advanced Optical Technologies, Erlangen, Germany, 3Siemens Healthcare, Erlangen, Germany, 4Siemens Medical Solutions, Malden, MA, United States, 5Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States
Synopsis
Estimating intrascan subject motion enables the reduction of motion artifacts but often requires further calibration data e.g. from an additional motion-free reference. In this work, we explore how the reuse of supplementary scans in the imaging workflow can be used as motion calibration data. More specifically, the preceding parallel imaging calibration scan is reutilized to support a Deep Learning (DL) approach for estimating motion. Results are presented which indicate that DL, in contrast to a conventional optimization approach, can extract the motion and improve the image quality despite contrast differences between the calibration and imaging scans.
Introduction
Many proposed methods for retrospective MRI motion correction estimate intrascan motion and the motion-corrected image1,2. Such alternating optimizations based on minimizing the data inconsistencies are computationally expensive and highly non-convex, in part because of a lack of an absolute reference image or orientation .Providing a motion-free reference does not only solves resolves this ambiguity, but more importantly, can reduce the optimization to one single variable. In SAMER3, a short so-called scout is acquired and serves as a reference image for the motion parameter estimation. We have shown in previous work4 that Deep Learning (DL) can be used for estimating motion parameters to a reference, as a replacement of the aforementioned conventional optimization approaches1,2. The estimated motion parameters can then be included in the image reconstruction. Both algorithms, SAMER and MoPED, were proposed with strict requirements to the reference, which necessitated an extra acquisition.
In this work we investigate whether these methods can tolerate a deviation from these requirements.
In a typical scan, calibration data are often quickly acquired just prior to the imaging data. Most prominently is the acquisition of a fully sampled, low-resolution parallel imaging calibration scan (“PATRef”) for calculating the GRAPPA weights or sensitivity maps5. This PATRef scan is optimized to be as short as possible to minimize its effect on the protocol duration; however, its contrast usually does not match the imaging scan.
To avoid additional overhead of acquiring a reference for the motion correction we investigated whether this PATRef scan could be reutilized as a reference in adapted versions of SAMER and MoPED.
Methods
We acquired 42 motion free measurements of heads and phantoms in axial, sagittal and coronal orientation with a Cartesian TSE (TR: 6100ms, TE: 103ms, FA: 150°, FOV: 220mm×220mm, resolution: 448×448, acceleration factor: 2, phase resolution: 80%), using various Siemens MAGNETOM scanners (Siemens Healthcare, Erlangen, Germany) at 3 T. This results in 28 slices per volume acquired in 9 equidistant echo trains (ET) per slice with an ET length of 20. The protocol was configured to acquire an external PATRef scan (TE: 71ms, ET FA: 20°, TA: 4s) with 38 lines in phase encode direction and 64 samples in read-out direction.The data was split in train, validation, and test sets; random in-plane motion was simulated for a supervised training. A network based on MoPED (Fig. 1, referred as “MoPED PATRef”) was trained to estimate the 9 ET x 3 (Tx, Ty and Rz) motion parameters as proposed previously4 but with the PATRef scan as reference. Prior experiments showed that the best performance is achieved with retrospectively undersampling the PATRef to match the sampled motion k-space lines. In contrast to the previously proposed MoPED with matching contrast (“original MoPED”), we achieved iterative refinement of the motion parameters by adapting the reference to the prior estimation for each ET. This avoids model errors and is only possible with the larger sampled PATRef scan.
For SAMER, the scout image that is acquired with a contrast similar to the imaging scan was replaced with the reconstruction of the PATRef scan.
Furthermore, we compare in simulations the performance of MoPED PATRef and SAMER with PATRef (“SAMER PATRef”) scan to the performance using the ideal reference (i.e. matching contrast; “original SAMER”), which is the best theoretically achievable result.
Finally, a subject was asked to perform a nodding-up movement during the acquisition to evaluate on in-vivo data. For the original MoPED a motion-free scan was performed before.
Results
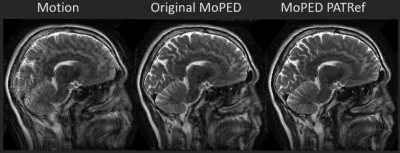
In Fig. 2 the regression plots show the estimate against the ground truth motion for the test set. It compares MoPED PATRef to the original MoPED. As can be seen, the results for MoPED PATRef exhibit a higher standard deviation than when using an exactly matched reference, but they are still in a very good agreement with the ground truth.Fig. 3 displays the motion correction results of simulations of the original MoPED, MoPED PATRef as well as original SAMER and SAMER PATRef. As expected, SAMER and MoPED original perform best; MoPED PATRef leads to an only minimally inferior image quality. For SAMER PATRef on the other hand the correction fails. Similar results are achieved with in-vivo data, shown in Fig. 4, where MoPED PATRef even leads to slightly better image quality.
Discussion and conclusion
The results show that our proposed DL based approach can learn how to handle contrast differences in the motion detection task. SAMER PATRef on the other hand was not able to converge, presumably due to the strong contrast difference. The optimization in SAMER relies on a contrast matched scout, which is accelerated by undersampling, whereas the PATRef scan is fully sampled but accelerated by selecting appropriate imaging parameters. MoPED on the other hand can deal with the different image appearance, leading to no additional scan time overhead. As shown, high improvements in image quality can be obtained, even in in-vivo acquisitions.In this scenario MoPED PATRef leads to only minimally degraded image quality compared to the optimal scenario of the original MoPED. Future work will have to investigate whether these results can be further improved by including more data sets into the training or by slight adaptions of the PATRef scan.
Acknowledgements
No acknowledgement found.References
1Haskell M., “Targeted motion estimation and reduction (TAMER): Data consistency based motion mitigation for MRI using a reduced model joint optimization,” IEEE Trans. Med. Imag, vol. 37, no. 5, pp. 1253-1265
2Cordero-Grande L., “Sensitivity encoding for aligned multishot magnetic resonance reconstruction,” IEEE Trans. Comput. Imag., vol. 2, no. 3, Sep. 2016.
3Polak D., “Scout accelerated motion estimation and reduction (SAMER),” Magn. Reson. Med., vol. 82, no. 4, pp. 1-16, Aug 2021.
4Hossbach J., “MoPED: Motion Parameter Estimation DenseNet for accelerating retrospective motion correction” ISMRM 2020 Nr. 3497
5Pruessmann K., “SENSE: Sensitivity Encoding for Fast MRI” Magn. Reson. Med. Vol. 42 pp. 952-962 1999
Figures