1890
Integrating hybrid CSI/EPSI acquisitions with L2 regularization for lipid removal of MRSI1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2The First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou, China, 3Renji Hospital Affiliated to Shanghai Jiaotong University School of Medicine, Shanghai Jiao Tong University, Shanghai, China
Synopsis
Placements of lipid saturation bands not only demand a skillful MRSI scanning operation but also restrict the full brain MRSI coverage. Therefore, MRSI acquisition without lipid suppression while doing post-removal of lipid becomes clinically attractive and valuable. In this work, we propose to integrate hybrid CSI/EPSI acquisitions with L2 regularization for fast lipid removal of MRSI. The hybrid CSI/EPSI acquisitions rely on the same excitation and refocusing evolution with the modified CSI and EPSI sequences. Phantom and in-vivo brain studies demonstrate the effectiveness and low computational cost of the proposed method.
Introduction
Magnetic resonance spectroscopic imaging (MRSI) enables spatial mapping of multiple tissue metabolites in vivo, many of which are proven to be valuable biomarkers for several diseases [1-2]. Chemical shift imaging (CSI) acquisition generally has a limited k-space coverage but high SNR while echo-planar spectroscopic imaging (EPSI) significantly accelerates MRSI and allows an extended k-space coverage with a low SNR within a short period [3]. Placements of lipid saturation bands not only demand a skillful MRSI scanning operation but also restrict the full brain MRSI coverage. Therefore, acquisition without lipid suppression while post-removal of lipid becomes clinically attractive. A union-of-subspaces (UOS) model [4] was proposed to remove nuisance signals in hybrid data with limited and sparse (k-t)-space coverage. However, the UOS algorithm requires accurate prior knowledge of spectral supports of lipid regions and takes a long reconstruction time. In this work, we propose to integrate hybrid CSI/EPSI acquisitions with L2 regularization for lipid removal of MRSI. Phantom and in-vivo brain studies demonstrate the effectiveness and low computational cost of the proposed method.Theory and methods
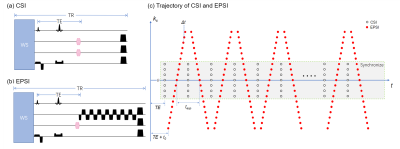
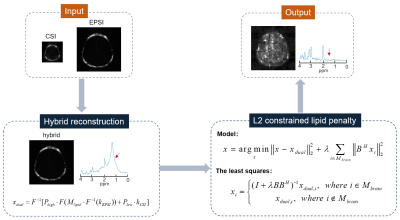
The hybrid CSI/EPSI acquisitions rely on the same excitation and refocusing evolution on the modified CSI and EPSI sequence as shown in Fig. 1. There is a small interval of starting acquisition times of CSI and EPSI sequences, and EPSI acquisition has a tilting kx-t trajectory instead of the equal-spaced chemical shift encoding. All those trajectory differences as shown in Fig. 1c are correctable by synchronizing the center CSI/EPSI acquisitions with time-shifting correction and re-gridding. The flowchart of the reconstruction is shown in Fig. 2. A hybrid dataset is generated by adding the consistent CSI/EPSI acquisitions and then followed by a lipid penalty with L2-regularization.All experiments were conducted on a 3.0T United Imaging uMR790 system (Shanghai, China) equipped with a 32-channel receiver head coil. The CSI data was acquired by a spin-echo spectroscopy sequence. The EPSI had the same FOV, slice thickness, and TR/TE as the CSI scan. A phantom of a water container surrounded by grapeseed oil was used to assess the matching quality of hybrid data. The CSI data was acquired with parameters as 160×160 mm2 FOV, 10 mm slice thickness, TR/TE = 1500/135 ms, 24×24 image matrix size, and 1000 Hz spectral width. The rest parameters of EPSI were: 128×128 image matrix size and echo spacing = 0.5 ms. In vivo brain data from a healthy volunteer after receiving informed consent was acquired with only water suppression to evaluate the effectiveness of lipid removal. For in vivo experiment, the parameters of CSI data were set as 200×200 mm2 FOV, 10 mm slice thickness, TR/TE = 1500/135 ms, 16×16 image matrix size, and 1389 Hz spectral width. The rest parameters of EPSI were: 80×80 image matrix size and echo spacing = 0.72 ms.
Results and Discussion
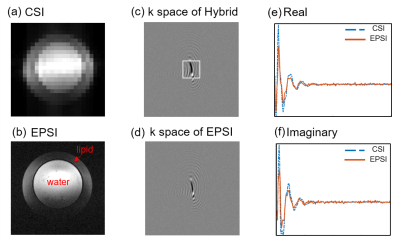
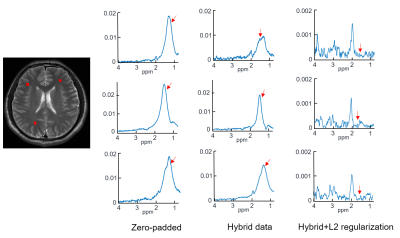
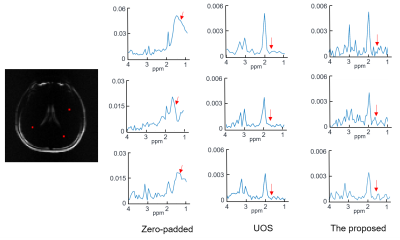
The quality of data matching between synchronized CSI and EPSI is shown in Fig. 3. The amplitude images of CSI and EPSI are shown in Fig. 3 (a) and (b). The grayscale images in Fig. 3 (c) and (d) correspond to the k-space data (real part) of the first time point of hybrid data and EPSI. The continuity in the hybrid data indicates a good match between the two types of data. The real and imaginary parts of the free induction decay signal from CSI and EPSI are illustrated in Fig. 3 (e) and (f). The close agreement of the profiles further reveals a good matching. The lipid removal results can be seen in Fig. 4. Severe leakages of the lipid signals are shown in the zero-padded CSI data (the first column). The lipid leakages were weakly attenuated in the hybrid data (the second column). No noticeable lipid signals were left in the last column with the processing of the proposed method. To further demonstrate the effectiveness of the proposed method, a comparison with the UOS is illustrated in Fig. 5. It can be seen that our method achieved comparable lipid suppression results with the UOS within 1 second, while the UOS takes about 35 minutes.Conclusion
We propose to integrate hybrid CSI/EPSI acquisitions with L2 regularization for lipid removal of MRSI. Phantom and in-vivo brain studies demonstrate the effectiveness and low computational cost of the proposed method.Acknowledgements
This work is supported by the National Science Foundation of China (No. 62001290) and Sponsored by Shanghai Sailing Program (20YF1420900).References
[1] Bilgic B, Chatnuntawech I, Fan A P, et al. Fast image reconstruction with L2‐regularization. Journal of magnetic resonance imaging, 2014, 40(1): 181-191.
[2] Bhattacharya I, Jacob M. Compartmentalized low‐rank recovery for high‐resolution lipid unsuppressed MRSI. Magnetic resonance in medicine, 2017, 78(4): 1267-1280.
[3] Metzger G, Sarkar S, Zhang X, et al. A hybrid technique for spectroscopic imaging with reduced truncation artifact. Magnetic resonance imaging, 1999, 17(3): 435-443.
[4] Ma C, Lam F, Johnson C L, et al. Removal of nuisance signals from limited and sparse 1H MRSI data using a union‐of‐subspaces model. Magnetic resonance in medicine, 2016, 75(2): 488-497.