1879
The Utility of Virtual Biopsies for Dataset Augmentation as Applied to AI-Based Detection of Tumor Infiltration in Non-Enhancing Brain Lesion1Graduate School, Medical College of Wisconsin, Wauwatosa, WI, United States, 2Biophysics, Medical College of Wisconsin, Wauwatosa, WI, United States
Synopsis
Delineation
of invasive tumor from peritumoral edematous tissue remains a major obstacle to
glioma treatment. To address this problem, a neural network was trained to
distinguish between these regions using biopsies paired with colocalized MRI
inputs. In addition to histologically confirmed biopsies, virtual biopsies
sampled from non-contrast enhancing, FLAIR enhancing regions of non-invasive
tumors (meningioma, metastasis) were used with an assumed classification of “non-tumor”.
The current work is a preliminary assessment of this assumption and it's impact
on model performance.
Introduction
Delineation of invasive tumor from peritumoral edematous tissue in non-contrast enhancing, FLAIR enhancing lesion (NEL) remains a major obstacle to glioma treatment [1,2]. In prior studies, our lab has successfully used biopsies extracted from NEL paired with colocalized MRI to train classification models to distinguish between these tissues [3-5].To account for significant class imbalance, non-tumor biopsies were virtually sampled from NEL of non-invasive tumors (meningiomas, metastases) without histological confirmation. Use of these “virtual biopsies” relies on the assumption that these samples are strongly correlated to NEL samples of invasive tumors. The current work is a preliminary assessment of this assumption and the impact these virtual biopsies have on model performance relative to models trained exclusively on histologically confirmed, or “true biopsies”.
Methods
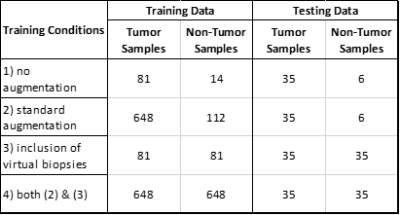
Dataset: Consented subjects with NEL biopsies, preoperative MRI (T1, T1+C, T2, FLAIR, DWI & DSC) and STEALTH imaging were selected from a brain tumor database (n=52). Bias correction and white matter normalization was applied to T1, T1+C, T2 & FLAIR. ADC maps (b=0,1000) were extracted from DWI and normalized, leakage-corrected rCBV and rCBF maps were extracted from DSC using Imaging Biometrics LLC software. Afni software was used to draw spherical ROIs on STEALTH imaging corresponding to biopsy extraction sites, linking histological assessment to precise regions on MRI. 116 tumor samples (WHO grades I-IV glial) and 20 non-tumor samples (primary glial, metastasis, meningioma) were collected this way for a total of 136 true biopsies. An additional 96 ROIs were drawn within NEL of the subjects with non-invasive tumor types to generate virtual biopsies.Model & Training: A neural network consisting of multiple 3D convolutional layers capped with fully connected layers was used for this assessment. Training was repeated under the following conditions: 1) no augmentations, 2) standard augmentations (flips, rotations), 3) inclusion of virtual biopsies, & 4) inclusion of both standard augmentations and virtual biopsies. Tumor and non-tumor data was split in a 70:30 ratio for training and testing groups, respectively. Note that the total number of samples varied depending on the which training conditions were used (shown in Table 1). Softmax cross entropy with L2 regularization was used for the loss function, RMSProp for the optimizer, and a grid search approach for hyperparameter selection. Training was done on a single Nvidia Tesla K40 gpu.
Results
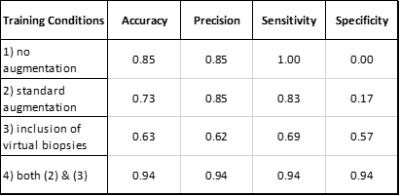
The accuracy, precision, sensitivity and specificity for the 4 different training conditions are given in Table 2. Training the neural network on an imbalanced dataset without augmentations resulted in a model that classifies all samples as tumor while incorporation of either standard augmentations or virtual biopsies resulted in more balanced models with varying success. The model trained with both standard augmentations and virtual biopsies performed well with .94 for each metric.Discussion/Conslusion
As expected, training a neural network with few samples of a given class generates a model incapable of recognizing said class. Interestingly, inclusion of virtual biopsies seems to have a benefit similar to that of using standard augmentation methods, specifically reducing the number of false positives by exposing the model to more non-tumor samples. In fact, the low specificity metric for the model trained only with standard augmentations indicates the virtual biopsies may better address the class imbalance problem.While these preliminary results are promising, interpretation is limited. An ideal assessment would evaluate model performance against a more balanced collection of true and virtual biopsies to ensure predictive capacity specifically improves with respect true, non-tumor biopsies because this is ultimately the clinically relevant distinction. As the dataset continues to grow, a more robust assessment will be made possible in future iterations of this study.
Overall, these results indicate classifiers can learn relevant information from virtual biopsies sampled from peritumoral regions of non-invasive tumors with an assumed "non-tumor" label for the purpose of distinguishing the invasive rim from peritumoral edema of invasive tumors. This reduces the need for histologically confirmed non-tumor biopsies of the same region.
Acknowledgements
Funding support provided by NIH/NCI U01 CA176110, NIH/NCI R01 CA255123.
Computational support provided by the Research Computing Center at Medical College of Wisconsin
FCOI: Imaging Biometrics LLC (KMS-financial interest), IQ-AI Ltd (KMS-ownership interest), Prism Clinical Imaging Inc (KMS-ownership interest)
References
1) Lasocki A, Gaillard F. Non-Contrast-Enhancing Tumor: A New Frontier in Glioblastoma Research. AJNR Am J Neuroradiol. 2019 May;40(5):758-765. doi: 10.3174/ajnr.A6025. Epub 2019 Apr 4. PMID: 30948373; PMCID: PMC7053910.
2) Eidel O, Burth S, Neumann JO, et al. Tumor infiltration in enhancing and non-enhancing parts of glioblastoma: a correlation with histopathology. PLoS One 2017;12:e0169292
3) Wujek R, Prah M, Al-Gizawiy M, Schmainda K. “Machine Learning Based Detection of Infiltrative Tumor within Non-Enhancing Peritumoral Regions” [abstract #2807]. In: Proceeding of the 59th Annual Meeting of the American Society of Neuroradiology; May 30-June 4 2020
4) Prah M, Wujek R, Al-Gizawiy M, Schmainda K. “Multi parameter MRI detection of infiltrative brain tumor validated with spatially-correlated brain tumor biopsies” [abstract #2723]. In: Proceeding of the 59th Annual Meeting of the American Society of Neuroradiology; May 30-June 4 2020
5) Wujek R, Prah M, Al-Gizawiy M, Schmainda K. “Machine Learning Based Classification of Tumor within Non-Enhancing Brain Lesions“ [abstract #1882]. In: Proceeding of the 2020 ISMRM & SMRT Virtual Conference & Exhibition; 08-14 August 2020
Figures