1877
A deep learning-basedquality control system for co-registration of prostate MR images
Mohammed R. S. Sunoqrot1, Kirsten M. Selnæs1,2, Bendik S. Abrahamsen1, Alexandros Patsanis1, Gabriel A. Nketiah1,2, Tone F. Bathen1,2, and Mattijs Elschot1,2
1Department of Circulation and Medical Imaging, Norwegian University of Science and Technology (NTNU), Trondheim, Norway, 2Department of Radiology and Nuclear Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway
1Department of Circulation and Medical Imaging, Norwegian University of Science and Technology (NTNU), Trondheim, Norway, 2Department of Radiology and Nuclear Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway
Synopsis
Multiparametric MRI (mpMRI) is a valuable tool for the diagnosis of prostate cancer. Computer-aided detection and diagnosis (CAD) systems have the potential to improve robustness and efficiency compared to traditional radiological reading of mpMRI in prostate cancer. Co-registration of diffusion-weighted and T2-weighted images is a crucial step of CAD but is not always flawless. Automated detection of poorly co-registered cases would therefore be a useful supplement. This work shows that a fully automated quality control system for co-registration of prostate MR images based on deep learning has potential for this purpose.
INTRODUCTION
Multiparametric MRI (mpMRI) is a valuable tool for the diagnosis of prostate cancer.1 However, conventional manual radiological reading requires expertise, is time-consuming, and is subject to inter-observer variability.2 To overcome these limitations, computer-aided diagnosis (CAD) systems have been proposed.2 mpMRI involves the acquisition and integration of T2-weighted imaging (T2W), diffusion-weighted imaging (DWI), and sometimes dynamic contrast-enhanced sequences.1 Image co-registration, in which two sequences are placed in the same spatial position and aligned, is an important step in the workflow of CAD.2,3 Co-registration allows the extraction of features from the same volume of interest using different sequences, which could improve the performance of the classification process and thus diagnosis by providing more representative quantitative information.3 In mpMRI of the prostate, it is common to co-register DWI to T2W images (T2WIs). However, manual quality control (QC) is currently still required, as poorly registered images may lead to poor CAD performance and thus inaccurate diagnosis. The aim of this work was to develop a fully automated QC system for co-registering prostate DWIs to T2WIs using a deep learning approach.METHODS
Datasets:The publicly available PROSTATEx challenge mpMRI dataset4 (n=346) was used. Cases with matching number of slices for transverse T2WIs and DWIs (b=50) were included (n=181) and the rest were excluded.
Workflow:
The pipeline of the proposed system (Figure 1) starts with 3D Image-to-Image translation5 of DWIs from T2WIs, to mimic perfectly co-registered DWIs. Then, translated DWIs (DWIt) are transformed to produce 3D deformed images (DWId) and a reference deformation score (RS) representing the maximum voxel displacement between modalities. From this, both the DWId and original 3D T2WIs were pre-processed by centre cropping (50%, 50%, and 80% for the X, Y, and Z axes, respectively), rescaling to the 99th percentile intensity value, and resizing the slices to 64×64 pixels with 15 slices and 0.5×0.5 mm in-plane resolution. Together with the RS labels, the pre-processed DWId and T2WIs were used as 3D, 2-channel input to a regression fully connected convolutional neural network (rfCNN). This network was trained to estimate the deformation score (ES). The processing, training, and analysis were performed with MATLAB R2021b (MathWorks, , USA) on a single NVIDIA Quadro T2000 16 GB GPU in Windows 10.
Image-to-Image translation was performed using a cycle-consistent generative adversarial network (CycleGAN).5 70% of the dataset was used for training, and all images were inferred. CycleGAN structure and hyperparameters are shown in Figure 2.
Image deformation was performed using tranformix6 (Elastix v4.7). DWIt were Euler transformed with 6 randomly generated rotation and translation parameters and resampled with a BSpline interpolator. Transformix generated a deformation field matrix, which was first centre cropped according to pre-processing, and then used to calculate the maximum voxel displacement as RS. Each of the 3D DWIt images was deformed 20 times to produce RSs between 0-25.5 mm. The resulting DWId dataset had a size of 3620 3D images with corresponding T2WI and RS.
rfCNN was trained (70%, same patients as CycleGAN training) and tested (30%) using this dataset, split at patient level. The network layers and hyperparameters are shown in Figure 4.
Performance evaluation:
CycleGAN performance was evaluated by visual inspection. Histograms were plotted for RSs distribution to ensure a realistic representation of the registration deformation in the rfCNN train and test sets. Wilcoxon-rank-sum test was used to compare the two distributions. Root-mean-square error (RMSE), mean absolute error (MAE), correlation plot, Bland-Altman plot, and Spearman's rank test between RSs and ESs were used to evaluate the performance of the QC system.
RESULTS
Figure 2A displays CycleGAN's trainingprogress. Visual inspection of randomly selected output cases showed acceptable
results, although typical upsampling artefacts tend to present as checkerboard patterns.
Figure 2B shows an example of two translated images. Figure 3A shows an example
of pre-processed DWId images and their corresponding pre-processed T2WIs and
RSs. The RSs distribution of the rfCNN train and test sets (Figure 3B) was similar
(p=0.07) and skewed toward lower values, mimicking the expected realistic co-registration
deformation. For the rfCNN test dataset, the RMSE, MAE±standard deviation, and
interquartile range were 2.71, 2.11±1.71, and 5.18 mm, respectively. The
correlation plot (Figure 5A) and Bland-Altman plot (Figure 5B) show acceptable
performance with a slight average overestimation (mean=1.39 mm).
DISCUSSION
Initial analysis of our proposed QC system showedacceptable results in terms of predicting the maximum voxel displacement of the
3D DWId images paired with T2WIs. The relatively wide 95% CI leave room for
improvement. The model shows a tendency to overestimate the RS, but this appears
to be systematic, which can be considered when applying the system. In
practice, this system should be adapted to the application in question,
defining a threshold to automatically distinguish between acceptable and poor co-registrations.
Image-to-Image translation was used to generate DWIs
that can be used as a reference for the development of the QC system, since the
original DWIs are not necessarily perfectly co-registered to the original
T2WIs. However, evaluation of the method’s performance on real instead of
generated DWIs data will be the next step towards clinical implementation.
CONCLUSION
We developed a fullyautomated system for quality controlling DWI to T2W co-registration, which
shows promising initial results for prostate imaging.
Acknowledgements
We would like to thank the organizers of the PROSTATEx challenge for making their datasets available.References
- Turkbey B, Rosenkrantz AB, Haider MA, et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur Urol. 2019;76(3):340-351.
- Lemaître G, Martí R, Freixenet J, et al. Computer-Aided Detection and diagnosis for prostate cancer based on mono and multi-parametric MRI- A review. Computers in Biology and Medicine. 2015;60(C):8–31.
- Song G, Han J, Zhao Y, etal. A Review on Medical Image Registration as an Optimization Problem. Curr Med Imaging Rev. 2017;13(3):274-283.
- Armato SG 3rd, Huisman H, Drukker K, et al. PROSTATEx Challenges for computerized classification of prostate lesions from multiparametric magnetic resonance images. J Med Imaging (Bellingham). 2018;5(4):044501.
- Zhu J, Taesung P, Phillip I, et al. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. IEEE ICCV in Venice. 2017;2242–51.
- Klein S, Staring M, Murphy K, et al. elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29(1):196-205.
Figures
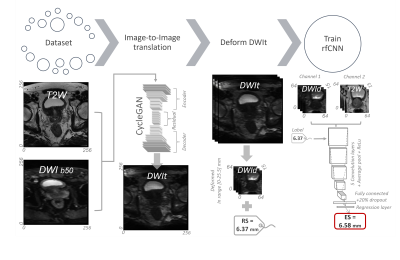
The pipeline of the proposed system. It starts with 3D image-to-image
translation of DWIs from T2WIs. Then, the translated DWIs (DWIt) are Euler
transformed to produce deformed images (DWId) and a reference deformation score
(RS). From this, both the DWId and the original 3D T2WIs were pre-processed to
size 64x64x15. The pre-processed DWId and T2WIs were then used as 3D 2-channel
input in addition to RS as a label for a regression fully connected
convolutional neural network (rfCNN) to predict an estimated deformation score
(ES).
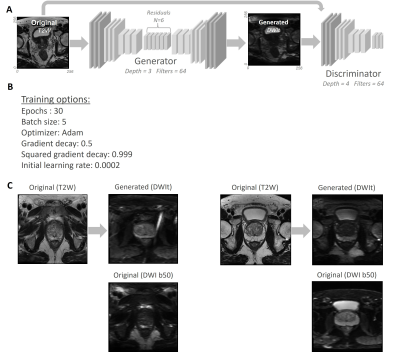
A) The structure of CycleGAN. B) The hyperparameters of CycleGAN. C) Two
examples of T2WIs and their corresponding generated DWIs using CycleGAN.
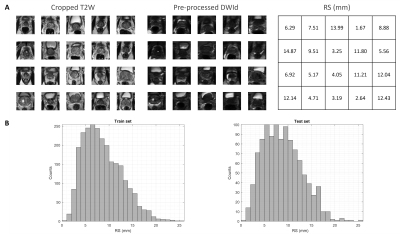
A) Example of pre-processed DWId images and their corresponding
pre-processed T2WIs and RSs.B) RSs distribution of the rfCNN train and test
sets.
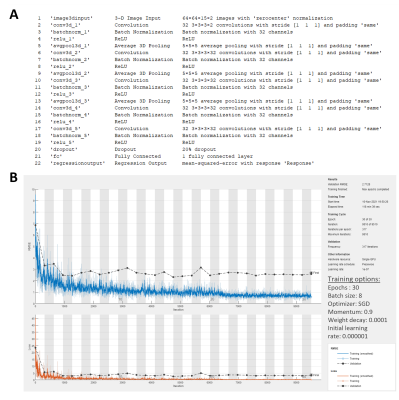
A) The structure and layers of rfCNN. B) The training progress plot,
which shows acceptable training performance, and hyperparameters of the rfCNN.
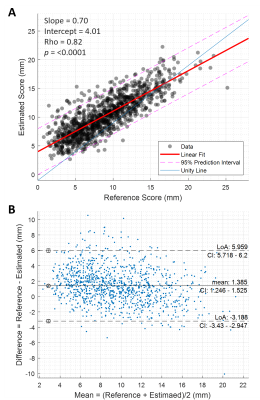
A) The correlation plot between the reference
and estimated deformation scores. B) The Bland-Altman plot for the reference
and estimated deformation scores.
DOI: https://doi.org/10.58530/2022/1877