1871
Impact Of MR Intensity Normalization For Different MR Sequences In MRI Based Radiomics Studies1CCU Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Medical Faculty, Heidelberg University Hospital, Heidelberg, Germany, 3Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg, Germany, 4German Cancer Consortium (DKTK) Core Center, Heidelberg, Germany, 5Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany, 6CCU Radiation Therapy, German Cancer Research Center (DKFZ), Heidelberg, Germany, 7National Center for Tumor Diseases (NCT), Heidelberg, Germany
Synopsis
MR-based radiomics prognostic signatures have great clinical potential but are currently hindered due to a lack of standardisation in the radiomics workflow. This study focuses on a crucial step of these workflows, i.e. intensity normalisation, while presenting a methodology that allows for determining the most suitable MR intensity normalization method for a specific entity.
Introduction
Robust Magnetic Resonance (MR) -based radiomics 1 models often require large amounts of data. Therefore, MR images are usually collected from multiple centres, sites, and scanners. However, since MR intensities are acquired in arbitrary units, images from different scanners are not directly comparable. While this intensity variation has little effect on the clinical diagnosis, in radiomics studies, the performance of subsequent processing steps as image registration and segmentation, as well as model-building, might be impacted. Thus, intensity normalization is essential. The image biomarker standardization initiative 2 (IBSI) has set definitions of a general standardized radiomics image processing workflow. However, no specific guidelines on the proper choice of intensity normalization methods are currently present. As the problem of reproducibility becomes more evident with the increase of published radiomics 3, more effort has been put into the detailed investigations of the effects of different image pre-processing steps on the overall robustness and reproducibility of the radiomics models 4-5. Our study aims to investigate the use of different normalization algorithms in multi-scanner brain MRI datasets, and more precisely, the performance of different methods on different sequences and their impact on the overall model's performance, by analyzing the predictive power of the methods' respective normalized dataset in the prediction of survival outcomes, i.e. overall survival (OS) and progression-free survival (PFS).Methods
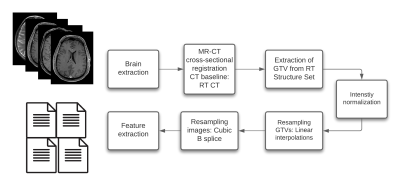
MR scans acquired before radiotherapy (RT) were retrieved from two independent data cohorts. The training cohort consists of 195 patients with pathologically confirmed recurrent high-grade glioma (rHGG), collected from 17 MR models and the testing cohort of 101 primary HGG (pHGG) patients collected from 11 MR models. The sequences studied are T1 weighted (w) pre-and post-contrast agent (CA), T2w, and fluid-attenuated inversion recovery (FLAIR). T1w images were first corrected for signal inhomogeneities using the N4 bias field correction algorithm. Then, all MR images were reoriented to a common orientation and skull-stripped. Furthermore, cross-sectional linear co-registrations of the T1w post- CA images were performed to the RT planning CT to generate the MR to CT affine matrix, which was applied to the Gross Tumor Volume (GTV) segmentations, extracted from the DICOM structure set (SS) objects, to bring the GTVs to the MR space. Next, intensity normalization was performed, where the algorithms considered were: Fuzzy C-Means (9 different masks combinations), kernel density estimation (KDE), gaussian mixture models (GMM) histogram matching-based (Nyúl and Udupa, NU), white-strip (WS), and z-score normalization. After Intensity normalization, the images and segmentations were re-sampled to a voxel size of 1x1x1 mm using a cubic spline and linear interpolation, respectively. The image processing diagram is shown in Fig1. Sequence-specific significant features (SF) from the GTV segmentations were then extracted from the training set using Pyradiomics6. In an attempt to neutralize the impact of grey-level discretization on the overall result, five different bin counts (bc) were implemented, resulting in five sets of features per normalization method. Sequence-specific significant features were then chosen based on a combination of 3 different feature selection methods (Lasso, Boruta, and univariate survival analysis feature selection under Cox proportional hazard). Survival analyses were conducted using Cox proportional hazard. A ranking score was then assigned based on the test set bc-averaged 10-fold cross-validated (CV) concordance index (C-I), the Akaike information criteria (AIC), and the average number of SF(nf) of both OS and PFS prediction models.Results
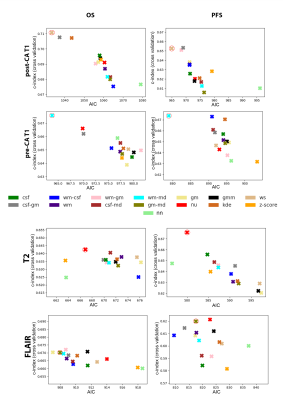
Scatter plots of the averaged 10-fold C-I against the AIC showed diversified results between the methods for each MR sequence (Fig.2). The WS and NU methods were ranked first for post-CA T1w (OS/PFS 10-fold CV C-I: 0.71/0.67, AIC: 1024/963, nf: 11/7) and T2w (0.64/0.66, 667/580, 4/6), respectively. FCM with a mask combination of the cerebrospinal fluid and mode ( (most common intensity value)) had the best performance for pre-CA T1w (0.69/0.68, 958/873, 7/7). As for FLAIR, FCM with a mask combination of the white matter and mode (0.67/0.61, 908/817, 5/5) was ranked first.Discussion
The MR intensity approach directly impacted the overall power of the radiomics based MR predictive models. Furthermore, considering the variability of the acquired results for the different MR sequences, it can be seen that the intensity normalization algorithm performance is correlated with the MR sequence and that the problem cannot be simplified to one intensity normalization technique. The methodology presented in this study can be further implemented to different entities to determine the most suitable MR intensity normalization algorithm.Conclusion
To summarize, variation in the results for the different MR sequences showed that the intensity normalization method performance is sequence-dependent and that it directly impacts the predictive power of survival models. Therefore, the documentation of the adapted normalization approach is necessary to enable the reproducibility of the MRI-based radiomics model.Acknowledgements
Funding by the H2020 MSCA-ITN PREDICT project.References
1. Lambin, P., Rios-Velazquez, E., Leijenaar, R., Carvalho, S., van Stiphout, R.G.P.M., Granton, P., Zegers, C.M.L., Gillies, R., Boellard, R., Dekker, A., Aerts, H.J.W.L.: Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 48, 441–446 (2012)
2. Zwanenburg, A. et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295, 328–338 (2020)
3. Ibrahim, A. et al. Radiomics for precision medicine: current challenges, future prospects, and the proposal of a new framework. Methods (2020)
4. Duron, L. et al. Gray-level discretization impacts reproducible MRI radiomics texture features. PloS one 14, e0213459(2019)
5. Carré, A. et al. Standardization of brain MR images across machines and protocols: bridging the gap for MRI-based radiomics. Sci. reports 10, 1–15 (2020)
6. Van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017)
Figures