1838
Intra- and intersubject synthesis of cardiac MR images using a VAE and GAN1Biomedical Engineering Department, Eindhoven University of Technology, Eindhoven, Netherlands, 2Philips Research Laboratories, Hamburg, Germany, 3MR R&D - Clinical Science, Philips Healthcare, Best, Netherlands
Synopsis
We propose a method for synthesizing cardiac MR images with plausible heart shape and realistic appearance. It breaks down the synthesis into labels deformation and label-to-image translation. The former is achieved via latent space interpolation in a VAE model, while the latter is accomplished via a conditional GAN model. We synthesize 32 short-axis slices within each subject (intrasubject), as well as eight intermediary generated subjects between two dissimilar real subjects (intersubject) that have different anatomies. Such method could provide a solution to enrich a database and to pave the way for the development of generalizable DL based algorithms.
Introduction
Deep generative modelling has gained attention in medical imaging research thanks to its ability to generate highly realistic images that may alleviate medical data scarcity [1]. The most successful family of generative models known as generative adversarial networks (GANs) [2] and Variational Autoencoders (VAEs) [3] are widely used for medical image synthesis and segmentation [4][5]. However, due to complex imaging features and anatomical characterises of cardiac MRI data generating plausible images of short-axis (SA) slices with accurate ground truth labels remains a challenge. Especially for the application of generating training labelled data that have varying appearance and anatomy for developing a robust deep-learning (DL) medical image analysis model.We propose to break down the task of image synthesis into 1) learning the deformation of anatomical content of the ground truth (GT) labels using VAEs and 2) translating GT labels to realistic CMR images using conditional GANs. We demonstrate intrasubject synthesis to generate intermediate SA slices within a given subject and intersubject synthesis to generate intermediate heart geometries between two different subjects acquired using different scanner vendors. The ultimate purpose of such synthesis would be to enrich the database of CMR images and provide accurate labels for a downstream supervised task, such as training a DL segmentation algorithm.
Methods
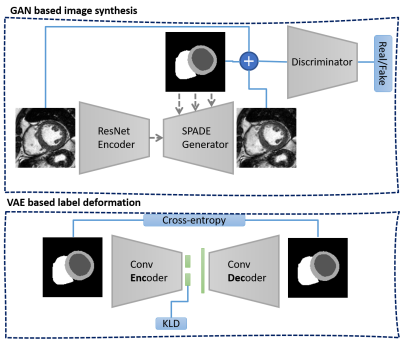
We utilize our conditional GAN model that employs a SPADE normalization layer throughout the generator architecture to preserve the content of the GT labels [6][7]. After successful training of the model with pairs of real images and corresponding labels, the generator can translate GT labels to realistic CMR images. To alter the heart anatomy of the synthesized image, we can simply deform the labels. Here we propose a DL based approach using a VAE to create plausible anatomical deformations of the heart labels which are subsequently utilized for synthesizing new subjects.A convolutional VAE model is designed and trained on GT labels of the heart to learn the distribution of different heart geometries presented in the database. After training, we encode each input label to its compressed embedding vector known as latent code. We linearly interpolate between two latent codes of provided input labels and feed them to the decoder network to reconstruct intermediate anatomical shapes. These newly deformed labels are used for image synthesis. The training and inference configuration of both models are shown in Figure 1 and Figure 2, respectively. For our experiments, we utilize CMR images from Siemens (vendor A) and Philips (vendor B) scanners provided by the M&Ms challenge [8]. All 150 subjects are resampled to 1.5 x 1.5 mm in-plane resolution and cropped to 128 x 128 pixels around the heart using the GT labels.
Experiments and Results
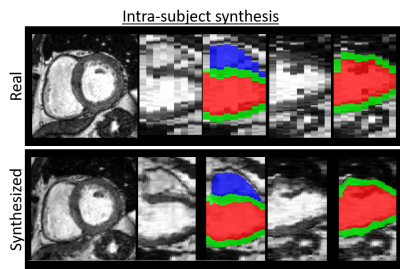
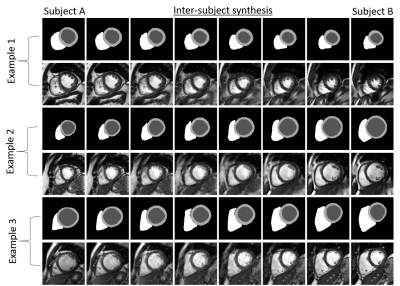
Experiments are designed for intra- and intersubject synthesis. For the intra-subject synthesis, we wish to increase the through-plane resolution of the SA slices for each subject. All slices are first encoded into the corresponding latent vectors followed by linear interpolation to increase to 32 latent vectors. These vectors are then reconstructed by the decoder network to create labels for intermediate slices. These labels are finally translated to images by the synthesis model, as depicted in Figure 3.Inter-subject synthesis aims to generate new examples with the intermediate heart anatomy between two dissimilar subjects. To this end, intra-subject synthesis is first performed to equalize the number of SA slices for each subject. Then following the same procedure, eight intermediate deformations are created by interpolating within the latent code corresponding to each subject. Finally, the deformed labels are used for synthesizing new subjects. Three examples for different levels of similarities between the heart geometries are shown in Figure 4. A smooth transition between subjects from vendor A and another from vendor B can be observed from the results.
Discussion and Conclusion
This study investigated an approach for realistic CMR image synthesis with plausible anatomies by separating the task into image generation using a conditional GAN and label deformation using a VAE model. Intra-subject synthesis was demonstrated to increase the through-plane resolution of SA images, while inter-subject synthesis was devised to generate subjects that have intermediary heart geometries and appearances.Such an approach could provide a solution to diversify and enrich the available database of cardiac MR images and to pave the way for the development of generalizable DL based image analysis algorithms. Further evaluation of the usability of the synthesized data is considered one of the future directions.
Acknowledgements
This research is a part of the OpenGTN project, supported by the European Union in the Marie Curie Innovative Training Networks (ITN) fellowship program under project No. 76446
References
[1] X. Yi, E. Walia, and P. Babyn, “Generative adversarial network in medical imaging: A review,” Med. Image Anal., vol. 58, p. 101552, Dec. 2019.
[2] I. J. Goodfellow et al., “Generative Adversarial Networks,” Commun. ACM, vol. 63, no. 11, pp. 139–144, Jun. 2014.
[3] D. P. Kingma and M. Welling, “Auto-encoding variational bayes,” in 2nd International Conference on Learning Representations, ICLR 2014 - Conference Track Proceedings, 2014.
[4] T. Joyce and S. Kozerke, “3D medical image synthesis by factorised representation and deformable model learning,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 2019, vol. 11827 LNCS, pp. 110–119.
[5] S. Kazeminia et al., “GANs for medical image analysis,” Artif. Intell. Med., vol. 109, p. 101938, Sep. 2020.
[6] T. Park, M. Y. Liu, T. C. Wang, and J. Y. Zhu, “Semantic image synthesis with spatially-adaptive normalization,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2019-June, pp. 2332–2341, Jun. 2019.
[7] Lustermans, D. R. P. R. M., Amirrajab, S., Veta, M., Breeuwer, M., & Scannell, C. M. (2021). Optimized Automated Cardiac MR Scar Quantification with GAN-Based Data Augmentation. arXiv, 2109(12940).
[8] V. M. Campello et al., "Multi-Centre, Multi-Vendor and Multi-Disease Cardiac Segmentation: The M&Ms Challenge," in IEEE Transactions on Medical Imaging, doi: 10.1109/TMI.2021.3090082.
Figures