1796
Deeping-Learning based acceleration for Serial MR imaging using Transformer (SMR-Transformer): Comparison with CNN on Three Datasets1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China, 2Xi’an Key Lab of Radiomics and Intelligent Perception, School of Information Sciences and Technology, Northwest University, Xi'an, China
Synopsis
Serial image acquisition is always required for quantitative and dynamic MRI, in which the spatial and temporal resolution as well as the contrast is restricted by acquisition speed. Herein we employed a novel transformer based deep-learning method for reconstruction of highly under-sampled MR serial images. The network architecture and input were designed to take advantages of Transformer’s capacity on global information learning along the redundant series dimension. SMR-transformer was compared with 3D Res-U-Net, on a self-acquired CEST dataset, a cardiac dataset from OCMR and a diffusion dataset from HCP. Both images and quantitative comparison indicated SMR-Transformer allowed fast serial MRI.
Introduction
For quantitative and dynamic MRI, a series of images is always needed to be acquired, while the image resolution and quality would be restricted by the scan time. Typical serial MR sequences include CEST images with multiple saturation offsets, diffusion-weighted images with multiple directions, dynamic imaging with multiple time points or temporal phases, and so on. Recently correlations among the serial images have been utilized for highly-accelerated acquisitions, using either compress sensing methods[1,2], or deep-learning approaches[2,3]. However, restricted by locality of convolution operation, traditional deep-learning techniques could not take full account of the correlation among serial direction and image dimensions.As a novel deep-learning framework, transformer has demonstrated great advantages in vision tasks owing to its learning ability of global information by modeling long-range dependency[5], like Swin-Unet[6]. Herein, we utilized the transformer’s global attention on serial contrast dimension, to reconstruct highly under-sampled MR serial images, which has been initially validated on three datasets of different application scenarios.Methods
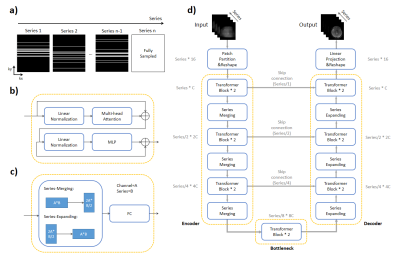
Under-sampling PatternFollowing theoretical hypothesis of compressed sensing, a variable density randomized cartesian under-sampling pattern was generated (Figure 1a), which is different for all series images to efficiently use the complementary information along the serial dimension. Notably, the last serial image was fully-sampled.
Dataset & Preprocessing
We demonstrate the performance on 3 datasets.
1)FastCEST (Fast acquired Chemical Exchange Saturation Transfer MR Dataset): A dataset containing 12 healthy subjects for chemical exchange saturation transfer imaging on 3T, acquired in the Center for Biomedical Imaging Research (CBIR), Tsinghua University, with written consent signed. For each subject, 10 slices and 31 different saturation offsets from -10 to 10ppm were acquired. A 10 times retrospective under-sampling was applied to all slices, with 10 subjects for training and 2 subjects for test.
2)OCMR (Open-Access Multi-Coil k-Space Dataset for Cardiovascular Magnetic Resonance Imaging): The cardiac cine data were interpolated to 32 frames and under-sampled by 8 times in k-space. 58 subjects were taken for training set and 8 subjects for test.
3)HCP (Human Connectome Project):10 subjects were selected for our study. Each case contains 40 slices on 288 gradient directions (including 18 B0 images). An 8 times under-sampling was applied to the k-space data after 2-dimensional Fast Fourier Transform.
Network Architecture
Figure 1 depicts the architecture of the proposed SMR-Transformer, which contains encoder and decoder sub-network like U-Net[7], but the convolution operations were replaced by transformer blocks. In the encoder sub-network, six transformer blocks followed by three series merging operations were performed for learning global contextual features. Symmetric to the encoder, three series-expanding operations followed by six transformer blocks were used to generate recovery feature maps for reconstruction. The transformer first learned global information along the series dimension through multi-head attention module, then encoded or decoded every series image through multi-layer perceptron (MLP) layer.
Network Training
The SMR-transformer network randomly picked the 4X4XNs patch as input(Ns is image number included in the series). Learning rate was set to 1e-4, then decayed by 10 times at 30 and 150 epochs. The number of training epochs were set as 300. Loss function was mean square error (MSE) and optimizer was Adam. For comparison, the regular Res-U-Net was also trained using NxXNyXNs patch size, in which Nx, Ny were chosen as the maximum number allowed by the GPU memory (FastCEST: 128X112X32, OCMR: 160X144X32, HCP:112X96X288).
Results and Discussion
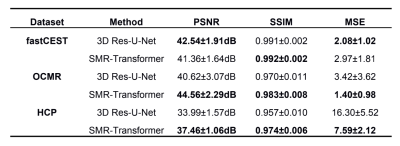
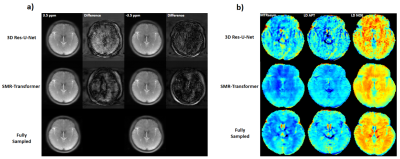
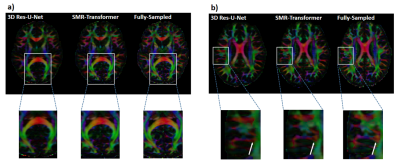
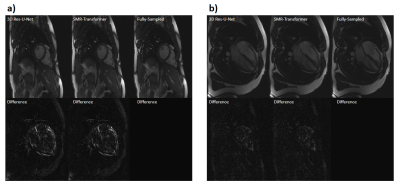
We evaluated the performance of proposed SMR-Transformer on three datasets, with the 3D Res-U-Net for comparison. Compared with Res-U-Net, our SMR-Transformer achieved higher PSNR and SSIM, and lower MSE on both the HCP and OCMR datasets; while for FastCEST dataset the two networks had similar PSNR and MSE (Table 1). For FastCEST, SMR-Transformer displayed less noisy contrast maps than Res-U-Net, including MTR asymmetry and Lorentzian difference at the amide and the NOE frequency (Figure 2). For HCP dataset, the colored fractional anisotropy (CFA) maps calculated by the software FSL showed that, SMR-Transformer reconstructed more detail information than Res-U-Net (Figure 3). For cardiac cining images of OCMR, SMR-Transformer and Res-U-Net both displayed well reconstruction on test images. However, both methods could not well estimate cardiac motion. A possible reason is that, cine imaging is more of a spatial-temporal imaging modality, that is different from the above two multi-contrast dataset. In future, network architecture of SMR-Transformer may need adjustment to give more attention on spatial dimension.Overall, SMR-Transformer outperformed Res-U-net for three kinds of serial images. The credits may be transformer’s advantage on directly global attention to series dimension, which overcomes the localized shortage of convolution operations. In current implementation, requires a fully-sampled serial image as a reference input. There are two reasons for that. (1) A high-quality image can help improving the reconstruction of overall series. (2) The spatial size of transformer inputs was only 4*4 now, therefore an unaliased image is needed to de-alias other images effectively.
Conclusion
We proposed a pure transformer network architecture for accelerating serial MR imaging and evaluated it on three datasets. Quantitative results of reconstruction performance, as well as the image visualization, demonstrated that our SMR-Transformer outperformed 3D Res-U-Net. Additionally, the proposed SMR-transformer method could enable the acceleration up to 10 times on different datasets, which is promising in acceleration of serial MR imaging.Acknowledgements
The authors acknowledged funding from National Natural Science Foundation of China (82071914). and the startup package from Tsinghua University to Dr. Song.References
[1] X. Chen, Y. Yang, X. Cai, D. A. Auger, C. H. Meyer, M. Salerno, et al., "Accelerated two-dimensional cine DENSE cardiovascular magnetic resonance using compressed sensing and parallel imaging," Journal of Cardiovascular Magnetic Resonance, vol. 18, Jun 14 2016.
[2] H. She, J. S. Greer, S. Zhang, B. Li, J. Keupp, A. J. Madhuranthakam, et al., "Accelerating chemical exchange saturation transfer MRI with parallel blind compressed sensing," Magnetic Resonance in Medicine, vol. 81, pp. 504-513, Jan 2019.
[3] J. Schlemper, J. Caballero, J. V. Hajnal, A. N. Price, and D. Rueckert, "A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction," Ieee Transactions on Medical Imaging, vol. 37, pp. 491-503, Feb 2018.
[4] A. Bustin, N. Fuin, R. M. Botnar, and C. Prieto, "From Compressed-Sensing to Artificial Intelligence-Based Cardiac MRI Reconstruction," Frontiers in Cardiovascular Medicine, vol. 7, Feb 25 2020.
[5] H. Liu, J. Lu, J. Feng, and J. Zhou, "Two-Stream Transformer Networks for Video-Based Face Alignment," Ieee Transactions on Pattern Analysis and Machine Intelligence, vol. 40, pp. 2546-2554, Nov 2018.
[6] H. Cao, Y. Wang, J. Chen, D. Jiang, X. Zhang, Q. Tian, et al. (2021, May 01, 2021). Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. arXiv:2105.05537. Available:https://ui.adsabs.harvard.edu/abs/2021arXiv210505537C
[7] S. Wang, Z. Ke, H. Cheng, S. Jia, L. Ying, H. Zheng, et al., "DIMENSION: Dynamic MR imaging with both k-space and spatial prior knowledge obtained via multi-supervised network training," NMR in biomedicine, pp. e4131-e4131, 2019-Sep-04 2019.
Figures