1789
Predicting Perfusion Augmentation Using Deep Learning without Vasodilators1Department of Radiology, Stanford University, Stanford, CA, United States
Synopsis
We present a deep learning technique to predict cerebral perfusion after vasodilation challenges. A 3D convolutional neural network (CNN)-based encoder-decoder architecture was constructed to transform ASL perfusion images acquired pre-vasodilation into post-vasodilation images using an improved attention-gated 3D U-Net. Results showed that the prediction and ground truth were not significantly different. This technique will enable a drug-free MR procedure to study the hemodynamic of patients with high risk cerebrovascular diseases.
Introduction
Cerebrovascular reactivity (CVR) is an important biomarker that can evaluate the risk of cerebrovascular diseases 1. It can be measured using a stress test whereby the cerebral blood flow (CBF) is elevated by a vasodilator, such as Diamox. The amount of CBF augmentation after vasodilation reflects CVR and can be measured using the pre- and post-vasodilation CBF data. Arterial spin labeling is a quantitative and non-invasive MR technique for CBF measurement and pseudo-continuous ASL with multiple post-labeling delays (PLDs) has demonstrated high accuracy in CVR measurement using the gold standard PET imaging as the reference 2. However, the administration of vasodilators can cause adverse side effects and may not be suitable for all patients. Recent development in deep learning has enabled the prediction of hemodynamic response to vasodilators in PET imaging 3. Here, we present a deep learning model to predict the outcome of vasodilation on CBF measured by multi-PLD PCASL on healthy subjects.Methods
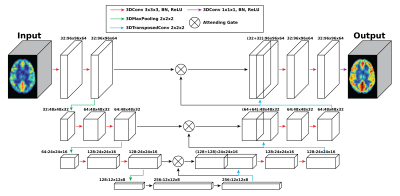
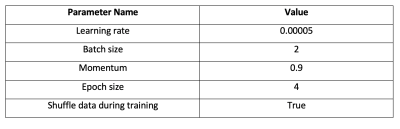
Multi-PLD PCASL data were collected from 64 normal subjects (23-57 years, 25 males) using a 3T GE simultaneous PET/MRI scanner. The main scanning parameters were: PLD = 300, 2000, 3700ms; labeling duration = 1700ms. The other scanning parameters of the multi-PLD PCASL were consistent with our previous work 2. A T1-weighted structural image was also acquired from each subject to facilitate brain extraction and registration. Diamox was administered to induce vasodilation during the scan at the dose of 15mg/kg of body weight of each subject with a maximum amount of 1000mg. ASL data were acquired before and 15 minutes after the injection of Diamox. Pre- and post-vasodilation CBF were computed by fitting the ASL difference data to the general kinetic model using BASIL 4. All CBF images were registered to the MNI-152 standard space for training and testing the deep learning model.An attention-based 3D convolutional encoder-decoder network was created to transform the pre-vasodilation CBF (input) into the post-vasodilation CBF (output), as shown in Figure 1 5. The training dataset included 54 pairs of pre- and post-vasodilation CBF and the testing dataset included 10 pairs. Data augmentation was employed to improve the robustness of the model using: (1) including both non-linearly and linearly registered CBF maps in standard space; (2) blurring the CBF maps with an FWHM=5mm filter; (3) applying a systematic bias of 5% increment in all CBF values to simulate the systematic bias in CBF measured by different ASL techniques. The hyperparameters of training the model were listed in Table 1. All training and testing were implemented using PyTorch 1.4 and performed on a Linux system with a GTX 1080 Ti GPU. To examine the performance of the trained model, post-vasodilation CBF was predicted using the pre-vasodilation CBF of the testing dataset. The prediction was compared with the ground truth using paired t-tests on a global (full brain) and regional basis.
Results
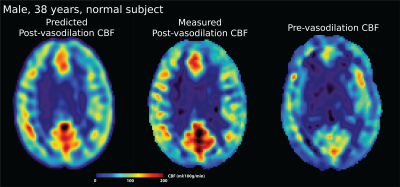
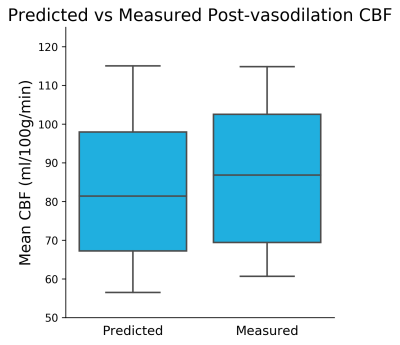
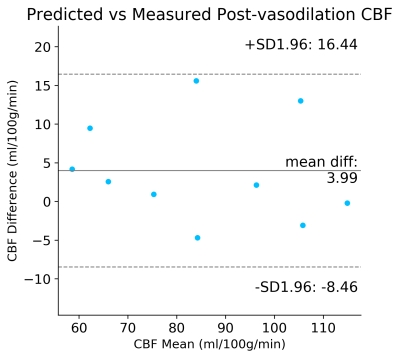
Figure 2 shows the predicted and measured post-vasodilation CBF maps and the pre-vasodilation CBF map. Figure 3 shows the mean CBF of the prediction and ground truth of the testing dataset. Figure 4 shows the Bland-Altman plot that compares the mean and difference of predicted and measured CBF. Overall, the results showed that no significant difference was found between the prediction and ground truth in the testing dataset.Discussion
Here, we developed a deep learning model to predict CBF augmentation without the need for vasodilators. Compared with the previous work that required multiple input images, the current model only required one CBF measured by multi-PLD PCASL, reducing the complexity of the model and reliance on high-resolution structural images. The model can be further improved by training on datasets of patients with pathology, which is an area of our ongoing investigation.Conclusions
Deep convolutional encoder-decoder networks can predict CBF augmentation accurately and potentially replace the need for vasodilators.Acknowledgements
This work is supported by the American Heart Association (Grant: 826254) and National Institutes of Health (Grant: R01EB025220-02 and P30 AG066515).References
1 Pinto J, Bright MG, Bulte DP, Figueiredo P. Cerebrovascular Reactivity Mapping Without Gas Challenges: A Methodological Guide. Front Physiol 2021; 11. doi:10.3389/fphys.2020.608475.
2 Zhao MY, Fan AP, Chen DY-T, Sokolska MJ, Guo J, Ishii Y et al. Cerebrovascular reactivity measurements using simultaneous 15O-water PET and ASL MRI: Impacts of arterial transit time, labeling efficiency, and hematocrit. NeuroImage 2021; 233: 117955.
3 Chen DYT, Ishii Y, Fan AP, Guo J, Zhao MY, Steinberg GK et al. Predicting PET Cerebrovascular Reserve with Deep Learning by Using Baseline MRI: A Pilot Investigation of a Drug-Free Brain Stress Test. Radiology 2020. doi:10.1148/radiol.2020192793.
4 Chappell MA, Groves AR, Whitcher B, Woolrich MW. Variational Bayesian Inference for a Nonlinear Forward Model. IEEE Transactions on Signal Processing 2009; 57: 223–236.
5 [1804.03999] Attention U-Net: Learning Where to Look for the Pancreas. https://arxiv.org/abs/1804.03999 (accessed 26 Sep2021).
Figures