1766
Improving U-Net based segmentation in cardiac parametrical T1 mapping by incorporating bounding box information1Charité Universitätsmedizin Berlin, Working Group on Cardiovascular Magnetic Resonance, Experimental and Clinical Research Center, a joint cooperation between the Charité Universitätsmedizin Berlin and the Max-Delbrück-Center for Molecular Medicine, Berlin, Germany, 2DZHK (German Centre for Cardiovascular Research), partner site Berlin, Berlin, Germany, 3Department of Cardiology and Nephrology, Helios Hospital Berlin-Buch, Berlin, Germany, 4Faculty for Computer Sciences, Hochschule Darmstadt (University of Applied Sciences), Darmstadt, Germany
Synopsis
Parametric mapping images contain to a large extent irrelevant background information. In order to hide some of it, we incorporated bounding box information into a U-net based segmentation network. Our dataset consisted of 845 training, 102 validation and 146 test T1 maps of native and post-contrast myocardium from different clinical studies, including healthy volunteers and patients with inflammatory heart disease, muscular dystrophies or chronic myocardial infarction. While cropping the image input improved the segmentation itself, a second input of the bounding box mask reduced the mean absolute and mean squared T1 deviation, which is clinically preferred.
Introduction
Parametric mapping is a quantitative technique for myocardial tissue characterisation that is considered one of the most important innovations in cardiovascular magnetic resonance imaging (CMR)1,2. While the conventional analysis of parametrical maps is time consuming due to manual segmentation, a recent study showed a novel fully automated segmentation pipeline3. This pipeline uses multiple independent U-Nets4 to predict a myocardial left ventricle (LV) segmentation mask based on the parametric map3. As the parametric mapping image contains irrelevant background information, we aimed to improve the prediction of an individual U-Net by incorporating bounding box information into the U-Net model.Methods
The parametric mapping dataset in this study consisted of native and post-contrast cardiac T1 maps from MOLLI sequences, that were measured on a 1.5T AvantoFit, 3.0T SkyraFit and 3.0T PrismaFit (all Siemens Healthineers, Erlangen, Germany). In total, 72 healthy volunteers (231 T1 maps) and 209 patients (862 T1 maps) with inflammatory heart disease, muscular dystrophies or chronic myocardial infarction from six studies were included. All images were manually segmented by an expert, who contoured the endo- and epicardial border of the myocardium excluding papillary muscles to determine the ground truth. For each of the six study sets, the data was randomly split with respect to number of subjects into 75% training (total=217 (845 T1 maps)), 10% validation (total=25 (102 T1 maps)) and 15% test (total=39 (146 T1 maps)) datasets.We compared the classical U-Net (refU) with a U-Net using an additional bounding box mask input (bbU) and a U-Net trained on bounding box cropped images (cropU). The bounding box was derived from another U-Net trained for bounding box prediction. In case of the cropU, the bounding box crop size was doubled in order to handle cases in which the predicted bounding box did not cover the entire LV region of interest. Figure 1 shows the schematic data flow of the three U-Nets described above.
All implemented U-Nets consisted of 27 layers and 6 skip connections. The model was trained on resized input images of 256x256 pixels using an Adam Optimizer with a clip norm of 0.001 and a learning rate of 0.001. The binary crossentropy served as the loss function. Training was set for a maximum of 1000 epochs, but was terminated early if the validation dice metric had not improved after a patience of 50 epochs. Additionally, a reduce on plateau learning rate scheme was set. This scheme halved the learning rate value if for 25 epochs the validation dice metric had no improvement. The used model learning rate, learning rate scheme and loss function were evaluated as optimal performing combination for all three U-Net models after comparing 30 different hyperparameter combinations.
In order to compare model performances, geometric metrics (dice similarity coefficient5 (DSC), average surface distance6 (ASD) and Hausdorff distance7 (HD)) and value based metrics (mean squared error (MSE) and mean absolute error (MAE)) were evaluated. Perfect agreement is reached, if DSC is maximized towards 100% and all other metrics converge towards zero. MSE and MAE are calculated with respect to the mean T1 value within segmentations. Significance was tested by both Friedman’s and Wilcoxon test with a significance level of 0.05.
Results
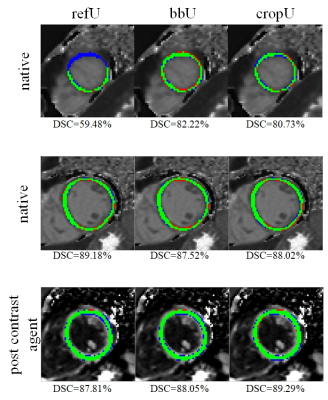
The cropU showed significantly better agreement with expert LV segmentation in terms of geometrics when compared with refU, but no significant improvements in MSE and MAE. In contrast, the bbU showed no significant difference with regards to the geometric metrices, but a significant advancement in MSE and MAE. The following results are averaged across the whole test dataset in the order of refU, bbU and cropU. Statistically significant differences with respect to refU are underlined. The geometric metrics of the test dataset read as follows: DSC (77.42% / 78.79% / 79.87%), ASD (2.84mm / 0.90mm / 0.80mm) and HD (6.32mm / 4.09mm / 3.13mm) while the value metrics were: MSE (1302.21ms2 / 305.26ms2 / 261.37ms2) and MAE (14.12ms / 10.70ms / 10.36ms). Figure 2 shows exemplary segmentation results of the three networks and the corresponding DSC.Discussion
While the automatic segmentation procedure is trained with respect to geometric similarity, in clinical routine a minimization of T1 value differences in the patient examination is crucial. The proposed additional bounding box feed-in into the U-Net showed an improvement in the geometrics, but no significant improvement in the value metrics for the cropU, while the bbU showed a significant improvement in the value metrics, but not in the geometry. For segmentation of parametric maps in a clinical context, bbU should be favoured over refU and cropU. Although the dataset covers different MR systems, slice positions, native and post contrast agent maps as well as healthy and diseased subjects, this study would benefit in a future extend from them being evenly distributed. Furthermore, a tighter cropping in the cropU might allow further geometric improvements to refine value metrics as well.Conclusion
Compared to a conventional U-Net, the incorporation of bounding box information in U-Net based segmentation either leads to improvements in geometric metrics (in case of cropU) or to improvements in value metrics (in case of bbU). Advancing in both would be ideally preferable.Acknowledgements
This study was supported by the BMBF (Bundesministerium für Bildung und Forschung) / DZHK (German Centre for Cardiovascular Research) via project FKZ81Z0100208 and complies with the declaration of Helsinki. The requirement for written informed consent was acquired during the original clinical studies and was therefore waived in this study due to its retrospective design.References
1. Messroghli DR, Moon JC, Ferreira VM, Grosse-Wortmann L, He T, Kellman P, Mascherbauer J, Nezafat R, Salerno M, Schelbert EB, Taylor AJ, Thompson R, Ugander M, van Heeswijk RB, Friedrich MG. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson. 2017;19:75.
2. Čelutkienė J, Plymen CM, Flachskampf FA, de Boer RA, Grapsa J, Manka R, Anderson L, Garbi M, Barberis V, Filardi PP, Gargiulo P, Zamorano JL, Lainscak M, Seferovic P, Ruschitzka F, Rosano GMC, Nihoyannopoulos P. Innovative imaging methods in heart failure: a shifting paradigm in cardiac assessment. Position statement on behalf of the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail. 2018;20:1615–1633.
3. Hann E, Popescu IA, Zhang Q, Gonzales RA, Barutçu A, Neubauer S, Ferreira VM, Piechnik SK. Deep neural network ensemble for on-the-fly quality control-driven segmentation of cardiac MRI T1 mapping. Med Image Anal. 2021;71:102029.
4. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. ArXiv150504597 Cs [Internet]. 2015 [cited 2021 Nov 9];Available from: http://arxiv.org/abs/1505.04597
5. Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26:297–302.
6. Ruskó L, Bekes G, Fidrich M. Automatic segmentation of the liver from multi- and single-phase contrast-enhanced CT images. Med Image Anal. 2009;13:871–882.
7. Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Mach Intell. 1993;15:850–863.
Figures