1761
Real-Time Spiral bSSFP Functional Cardiac MRI on a 0.55T Scanner Using a Deep Image Prior Reconstruction with GROG
Jesse Ian Hamilton1,2 and Nicole Seiberlich1,2
1Radiology, University of Michigan, Ann Arbor, MI, United States, 2Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
1Radiology, University of Michigan, Ann Arbor, MI, United States, 2Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Synopsis
This work introduces a physics-based deep learning reconstruction that can enable real-time (free-breathing and ungated) spiral 2D bSSFP functional cardiac MRI on a 0.55T scanner. The proposed method extends the concept of a deep image prior and does not require prior training on reference data. In addition, GROG is used to reduce the computational complexity of the forward model calculation by shifting spiral k-space data to their nearest Cartesian grid points, allowing use of FFT rather than NUFFT operations. Results are demonstrated in healthy subjects with functional measurements compared to gold standard acquisitions at 1.5T.
Introduction
Balanced steady state free precession (bSSFP) cine imaging is the gold standard for evaluating ventricular function1, and real-time techniques are useful in patients who cannot perform breathholds or do not have a regular cardiac rhythm. Low field scanners offer several potential advantages for bSSFP sequences; improved field homogeneity can mitigate banding artifacts, allowing use of efficient sampling methods with longer SNR-efficient readouts, and decreased SAR can permit higher flip angles to improve blood-myocardium contrast2–5. However, the globally decreased SNR, lower receiver coil counts (with suboptimal coil geometry), and lack of ECG gating capacity on lower-cost low field systems can present challenges for traditional cine imaging. Recently, deep learning has been applied for accelerated cine MRI6,7, including self-supervised approaches that do not require external training data8,9, which can be difficult to obtain for real-time applications. This work introduces a method for real-time spiral bSSFP functional cardiac imaging at 0.55T using a self-supervised deep image prior (DIP) reconstruction.Methods
The proposed reconstruction modifies the original deep image prior (DIP) technique10, using a U-net to generate real-time images from highly accelerated spiral data (Figure 1a). A tensor of uniformly distributed random numbers is input to the network and remains fixed during all steps. The U-net contains 2D convolutional layers with 5 downsampling/upsampling paths and skip connections, which places a smoothness prior on the images. To avoid time-consuming NUFFT operations11, the spiral k-space data are shifted to their nearest Cartesian grid points using GRAPPA Operator Gridding (GROG) before training12. The network is trained in a self-supervised manner (Figure 1b). The MRI encoding model is simulated, including multiplication by coil sensitivity maps, FFT, multiplication k-space undersampling masks, inverse FFT, and coil combination. This process yields an estimate of the aliased images that are compared to the acquired aliased images using a normalized l1-l2 loss, and the U-net is updated using backpropagation. In all experiments, training was performed with 1000 epochs, a mini-batch of 5 image frames, Adam optimizer (learning rate 0.001), and 10% dropout, implemented in TensorFlow 2.6.0 with Keras on a single GPU.The proposed DIP reconstruction was evaluated in simulations using the XCAT phantom13 with cardiac (60 bpm) and respiratory motion (12 breaths/second). Real-time scans were simulated using a 128x128 matrix with golden angle14 spiral sampling (48 interleaves to fully sample k-space) with acceleration factors of 3, 4, 6, 8, 12, 16, and 24. Images were reconstructed using zero-filling, l1-ESPIRiT with spatial (λ1=0.002 relative to maximum intensity) and temporal (λ2=0.01) total variation15, GRASP-Pro (64x64 for the low-resolution reconstruction and rank 20)16, and the DIP method. Reconstructions were compared to ground truth images using structural similarity (SSIM) averaged over all frames.
Next, data were acquired in nine healthy subjects on a 0.55T scanner (Free.Max, Siemens Healthineers) at a mid-ventricular slice using a bSSFP acquisition with 128x128 matrix, 300x300mm2 FOV, FA 110°, TR 6.3ms, and scan time 6.4s. A golden angle spiral trajectory was used with readout duration 3.8ms, Gmax 23.1 mT/m, and slewmax 34 mT/m/s. Eight spiral interleaves were grouped per frame (R=6) for a temporal resolution of 50.4ms. Images were reconstructed using zero-filling l1-ESPIRiT, GRASP-Pro, and DIP. Single-slice ejection fraction (EF) was measured after manual contouring of the myocardium. In four subjects, real-time scans at 0.55T were collected with full short-axis coverage of the left ventricle, and LVEF was compared to reference measurements at 1.5T (Sola, Siemens Healthineers) using a Cartesian breathheld ECG-gated bSSFP scan (FA 55-70°, 25 phases, TR/TE 2.4/1.2ms).
Results
In simulations (Figure 2), SSIM decreased with increasing acceleration factor for all methods. DIP yielded the smallest errors in all cases, and the difference was greater at higher acceleration factors. Figure 3 shows representative images from two subjects at 0.55T. Noise enhancement was observed on l1-ESPIRiT images; some temporal blurring was visible on GRASP-Pro images (e.g., papillary muscles during end-systole); DIP reduced noise and aliasing artifacts while preserving fine anatomical details. Figure 4 shows apical, medial, and basal real-time images in one subject at 0.55T reconstructed using DIP. Real-time images at 0.55T using all reconstructions yielded consistent single-slice EF (Table 1a); no significant differences were observed between DIP and either l1-ESPIRiT or GRASP-Pro using a paired t-test. LVEF calculated over all slices using real-time spiral imaging with DIP at 0.55T was in excellent agreement with gold standard acquisitions at 1.5T (Table 1b).Discussion
A physics-based deep learning reconstruction for real-time non-Cartesian MRI was introduced for free-breathing ungated functional cardiac imaging at 0.55T. This technique does not require prior training on reference data and instead employs self-supervised training. Related approaches for cine MRI based on the deep image prior framework have been proposed8,9. However, the technique described here does not require hand-crafted priors; assumptions about temporal dynamics (e.g., periodicity), which is important for free-breathing imaging; or additional regularization. This study also introduced a strategy for reducing the computational complexity of the forward model by using GROG, allowing use of the FFT rather than slower NUFFT operations.Conclusion
Real-time bSSFP functional cardiac imaging can be performed at 0.55T using golden angle spiral sampling with a GROG-facilitated deep image prior reconstruction that does not require prior training.Acknowledgements
NIH R01HL146021; Siemens Healthineers (Erlangen, Germany)References
- Bogaert J, Dymarkowski S, Taylor AM, Muthurangu V. Clinical Cardiac MRI. 2nd ed. Springer-Verlag Berlin Heidelberg; 2012.
- Campbell-Washburn AE, Ramasawmy R, Restivo MC, et al. Opportunities in interventional and diagnostic imaging by using high-performance low-field-strength MRI. Radiology. 2019;293(2):384-393.
- Bandettini WP, Shanbhag SM, Mancini C, et al. A comparison of cine CMR imaging at 0.55 T and 1.5 T. J Cardiovasc Magn Reson. 2020;22(1):37.
- Simonetti OP, Ahmad R. Low-Field Cardiac Magnetic Resonance Imaging: A Compelling Case for Cardiac Magnetic Resonance’s Future. Circ Cardiovasc Imaging. 2017;10(6):e005446.
- Srinivasan S, Ennis DB. Optimal flip angle for high contrast balanced SSFP cardiac cine imaging. Magn Reson Med. 2015;73(3):1095-1103.
- Sandino CM, Lai P, Vasanawala SS, Cheng JY. Accelerating cardiac cine MRI using a deep learning-based ESPIRiT reconstruction. Magn Reson Med. 2021;85(1):152-167.
- Biswas S, Aggarwal HK, Jacob M. Dynamic MRI using model-based deep learning and SToRM priors: MoDL-SToRM. Magn Reson Med. 2019;82(1):485-494.
- Yoo J, Jin KH, Gupta H, Yerly J, Stuber M, Unser M. Time-Dependent Deep Image Prior for Dynamic MRI. IEEE Trans Med Imaging. 2021:1. Early View. doi:10.1109/TMI.2021.3084288.
- Zou Q, Ahmed AH, Nagpal P, Kruger S, Jacob M. Dynamic Imaging Using a Deep Generative SToRM (Gen-SToRM) Model. IEEE Trans Med Imaging. 2021;40(11):3102-3112.
- Lempitsky V, Vedaldi A, Ulyanov D. Deep Image Prior. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE Computer Society; 2018:9446-9454.
- Fessler J, Sutton B. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Trans Signal Process. 2003;51(2):560-574.
- Seiberlich N, Breuer FA, Blaimer M, Barkauskas K, Jakob PM, Griswold MA. Non-Cartesian data reconstruction using GRAPPA operator gridding (GROG). Magn Reson Med. 2007;58(6):1257-1265.
- Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BMW. 4D XCAT phantom for multimodality imaging research. Med Phys. 2010;37(9):4902-4915.
- Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An optimal radial profile order based on the Golden Ratio for time-resolved MRI. IEEE Trans Med Imaging. 2007;26(1):68-76.
- Uecker M, Lai P, Murphy MJ, et al. ESPIRiT - An eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn Reson Med. 2014;71(3):990-1001.
- Feng L, Wen Q, Huang C, Tong A, Liu F, Chandarana H. GRASP-Pro: imProving GRASP DCE-MRI through self-calibrating subspace-modeling and contrast phase automation. Magn Reson Med. 2020;83(1):94-108.
Figures
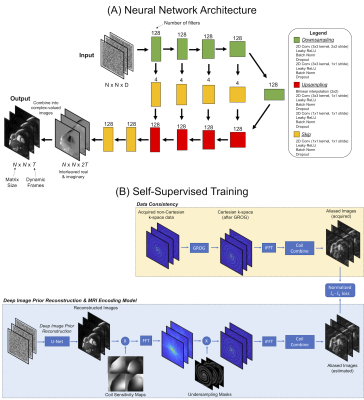
Figure 1: Deep image prior reconstruction for real-time non-Cartesian functional cardiac MRI. (a) Images are generated by a U-net with 5 downsampling/upsampling paths and skip connections. (b) During training, images output by the U-net are passed through the MRI encoding model, including multiplication by coil sensitivities, FFT, k-space undersampling, inverse FFT, and coil combination; the loss is calculated in the image domain. Before training, GROG is used to shift the spiral k-space data onto a Cartesian grid to avoid time-consuming NUFFT operations.
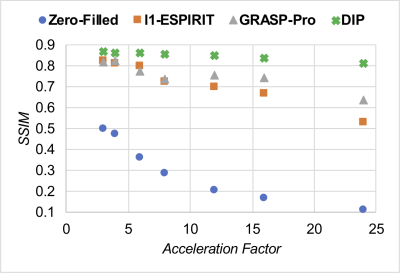
Figure 2: Simulations of free-breathing ungated spiral bSSFP functional cardiac scans using the XCAT phantom with different acceleration factors. Images were reconstructed using zero-filling, l1-ESPIRiT, GRASP-Pro, and the proposed deep image prior (DIP) reconstruction with GROG. Performance was quantified using structural similarity (SSIM).

Figure 3: Real-time spiral bSSFP functional cardiac images acquired at 0.55T in two healthy subjects. Images were reconstructed using zero-filling, l1-ESPIRiT, GRASP-Pro, and the proposed deep image prior (DIP) method. Relevant scan parameters include: 128x128 matrix, 300x300mm2 FOV, 110° flip angle, TR 6.3ms, total acquisition time 6.4 seconds, 128 frames, temporal resolution 50.4ms (8 spiral interleaves per frame, R=6).
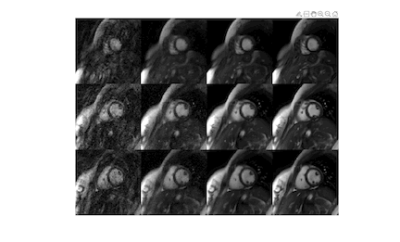
Figure 4: Real-time spiral bSSFP functional cardiac images at 0.55T at apical, medial, and basal slice positions acquired in a healthy subject. Images were reconstructed (from left to right) using zero-filling, l1-ESPIRiT, GRASP-Pro, and DIP. Scan parameters: 128x128 matrix, 300x300mm2 FOV, 50.4ms temporal resolution (8 spiral interleaves per frame, R=6), 110° flip angle, TR 6.3ms.
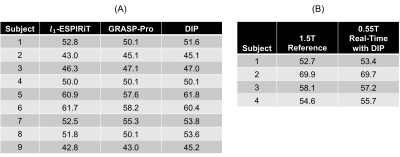
Table 1: Ejection fraction (EF) measurements. (a) Single-slice EF values are reported for 9 healthy subjects scanned at 0.55T using real-time spiral bSSFP functional cardiac imaging reconstructed with l1-ESPIRiT, GRASP-Pro, and the proposed deep image prior (DIP) technique with GROG. (b) In 4 subjects, real-time images at 0.55T were acquired with full coverage over the left ventricle. The table compares LVEF measured at 0.55T using the proposed technique with values obtained at 1.5T using a conventional Cartesian breathheld and ECG-gated scan.
DOI: https://doi.org/10.58530/2022/1761